This is a preprint.
Amino acid competition shapes Acinetobacter baumannii gut carriage
- PMID: 39502362
- PMCID: PMC11537318
- DOI: 10.1101/2024.10.19.619093
Amino acid competition shapes Acinetobacter baumannii gut carriage
Update in
-
Amino acid competition shapes Acinetobacter baumannii gut carriage.Cell Host Microbe. 2025 Aug 13;33(8):1396-1411.e9. doi: 10.1016/j.chom.2025.07.003. Epub 2025 Aug 4. Cell Host Microbe. 2025. PMID: 40763731 Free PMC article.
Abstract
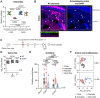
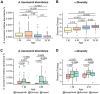
Antimicrobial resistance is an urgent threat to human health. Asymptomatic colonization is often critical for persistence of antimicrobial-resistant pathogens. Gut colonization by the antimicrobial-resistant priority pathogen Acinetobacter baumannii is associated with increased risk of clinical infection. Ecological factors shaping A. baumannii gut colonization remain unclear. Here we show that A. baumannii and other pathogenic Acinetobacter evolved to utilize the amino acid ornithine, a non-preferred carbon source. A. baumannii utilizes ornithine to compete with the resident microbiota and persist in the gut in mice. Supplemental dietary ornithine promotes long-term fecal shedding of A. baumannii. By contrast, supplementation of a preferred carbon source-monosodium glutamate (MSG)-abolishes the requirement for A. baumannii ornithine catabolism. Additionally, we report evidence for diet promoting A. baumannii gut carriage in humans. Together, these results highlight that evolution of ornithine catabolism allows A. baumannii to compete with the microbiota in the gut, a reservoir for pathogen spread.
Keywords: Acinetobacter baumannii; carbon preference; dietary supplements; gut colonization; microbiota; ornithine.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- World Health Organization (2024). WHO Bacterial Priority Pathogens List, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. (World Health Organization; ).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
