AP001885.4 promotes the proliferation of esophageal squamous cell carcinoma cells by histone lactylation- and NF-κB (p65)-dependent transcription activation and METTL3-mediated mRNA stability of c-myc
- PMID: 39502790
- PMCID: PMC11536669
- DOI: 10.1080/19768354.2024.2417458
AP001885.4 promotes the proliferation of esophageal squamous cell carcinoma cells by histone lactylation- and NF-κB (p65)-dependent transcription activation and METTL3-mediated mRNA stability of c-myc
Abstract
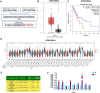
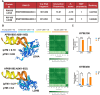
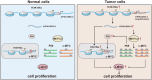
Esophageal squamous cell carcinoma (ESCC) is an aggressive malignant neoplasm, and up to now, the role of long non-coding RNA (lncRNA) AP001885.4 in cancer, including ESCC, is absolutely unclear. The GEPIA database was applied to identify differentially expressed and prognosis-associated genes in esophageal cancer (ESCA). CCK-8, colony formation, Western blot, and qRT-PCR methods were harnessed to investigate the role and mechanism of AP001885.4 in esophageal carcinogenesis. By analyzing TCGA data in the GEPIA database, two lncRNAs were selected. AP001885.4 was overexpressed and positively associated with the unfavorable outcome of ESCC patients, and LINC001786 was under-expressed and negatively linked with the poor prognosis. Knockdown of AP001885.4 suppressed the proliferation and colony formation of ESCC cells. Importantly, the silence of AP001885.4 downregulated c-myc. Mechanically, the knockdown of AP001885.4 reduced METTL3 expression and m6A modification in c-myc mRNA, and METTL3 positively regulated c-myc. Furthermore, the knockdown of AP001885.4 diminished histone lactylation and NF-κB (p65) expression, and the protein lactylation inhibitors (2-DG, 2-deoxy-D-glucose and oxamate) and the NF-κB inhibitor (JSH-23) also lessened c-myc expression. Consequently, our findings suggested that AP001885.4 promoted the proliferation of esophageal squamous cell carcinoma cells by histone lactylation- and NF-κB (p65)-dependent transcription activation and METTL3-mediated mRNA stability of c-myc.
Keywords: AP001885.4; C-myc; ESCC; NF-κB; lactylation.
© 2024 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
