Pneumococcal extracellular vesicles mediate horizontal gene transfer via the transformation machinery
- PMID: 39503503
- PMCID: PMC11656791
- DOI: 10.1128/msphere.00727-24
Pneumococcal extracellular vesicles mediate horizontal gene transfer via the transformation machinery
Abstract
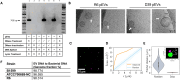
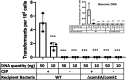
Bacterial cells secrete extracellular vesicles (EVs), the function of which is a matter of intense investigation. Here, we show that the EVs secreted by the human pathogen Streptococcus pneumoniae (pneumococcus) are associated with bacterial DNA on their surface and can deliver this DNA to the transformation machinery of competent cells. These findings suggest that EVs contribute to gene transfer in Gram-positive bacteria and, in doing so, may promote the spread of drug resistance genes in the population.IMPORTANCEThis work extends our understanding of horizontal gene transfer and the roles of extracellular vesicles in pneumococcus. This bacterium serves as the model for transformation, a process by which bacteria can take up naked DNA from the environment. Here, we show that extracellular vesicles secreted by the pneumococcus have DNA on their surface and that this DNA can be imported by the transformation machinery, facilitating gene transfer. Understanding EV-mediated gene transfer may provide new avenues to manage the spread of antibiotic drug resistance.
Keywords: Streptococcus pneumoniae; antibiotic resistance; competence; extracellular vesicle; horizontal gene transfer; transformation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Update of
-
Pneumococcal Extracellular Vesicles Mediate Horizontal Gene Transfer via the Transformation Machinery.bioRxiv [Preprint]. 2023 Dec 15:2023.12.15.571797. doi: 10.1101/2023.12.15.571797. bioRxiv. 2023. Update in: mSphere. 2024 Dec 19;9(12):e0072724. doi: 10.1128/msphere.00727-24. PMID: 38168155 Free PMC article. Updated. Preprint.
References
-
- Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, Covacci A, Mitchell TJ, Bentley SD, Kilian M, Ehrlich GD, Rappuoli R, Moxon ER, Masignani V. 2010. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol 11:R107. doi: 10.1186/gb-2010-11-10-r107 - DOI - PMC - PubMed
-
- Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. 2007. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol 189:8186–8195. doi: 10.1128/JB.00690-07 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
