Triptolide induces hepatotoxicity by promoting ferroptosis through Nrf2 degradation
- PMID: 39503881
- PMCID: PMC11541276
- DOI: 10.1007/s10565-024-09930-0
Triptolide induces hepatotoxicity by promoting ferroptosis through Nrf2 degradation
Abstract
Background: Triptolide (TP), a principal active substance from Tripterygium wilfordii, exhibits various pharmacological effects. However, its potential hepatotoxicity has always been a significant concern in clinical applications.
Purpose: This research aimed to explore the involvement of ferroptosis in TP-mediated hepatic injury and the underlying mechanisms.
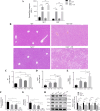
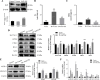
Methods: In this study, in vitro and in vivo experiments were involved. Hepatocyte damage caused by TP was evaluated using MTT assays, liver enzyme measurement and H&E staining technique. Ferroptosis was assessed by measuring iron level, lipid peroxide, glutathione (GSH), mitochondrial morphology and the key protein/mRNA expression implicated in ferroptosis. To verify the contribution of ferroptosis to TP-induced liver damage, the ferroptosis inhibitor Ferrostatin-1 (Fer-1) and a plasmid for overexpressing glutathione peroxidase 4 (GPX4) were employed. Subsequently, nuclear factor erythroid 2-related factor 2 (Nrf2) knockout mice and Nrf2 overexpression plasmid were utilized to investigate the underlying mechanisms. Nontargeted lipidomics was used to analyze lipid metabolism in mouse liver. Moreover, the cellular thermal shift assay (CETSA), cycloheximide (CHX) and MG132 treatments, and immunoprecipitation (IP) assays were applied to validate the binding of TP to Nrf2 and their interactions.
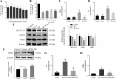
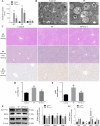
Results: TP triggered ferroptosis in hepatocytes, as indicated by iron accumulation and lipid peroxidation. Ferroptosis was responsible for TP-induced hepatic injury. During the process of TP-induced liver damage, the Nrf2 signaling pathway was significantly suppressed. Notably, the deletion of Nrf2 in mice aggravated the extent of liver injury and ferroptosis associated with TP, whereas enhancing Nrf2 expression in cells significantly reduced TP-induced ferroptosis. Additionally, dysregulation of lipid metabolism was associated with TP-induced liver injury. TP may directly bind to Nrf2 and enhance its degradation through the ubiquitin-proteasome pathway, thereby inhibiting or reducing Nrf2 expression.
Conclusion: In summary, the suppression of Nrf2 by TP facilitated the occurrence of ferroptosis, resulting in liver damage.
Keywords: Ferroptosis; Liver injury; Nrf2; Triptolide.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Aldosary BM, Sutter ME, Schwartz M, Morgan BW. Case series of selenium toxicity from a nutritional supplement. Clin Toxicol (Phila). 2012;50:57–64. - PubMed
-
- Andrade RJ, Chalasani N, Bjornsson ES, Suzuki A, Kullak-Ublick GA, Watkins PB, Devarbhavi H, Merz M, Lucena MI, Kaplowitz N, Aithal GP. Drug-induced liver injury. Nat Rev Dis Primers. 2019;5:58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2021JJ30932/Natural Science Foundation of Hunan Province
- 2020JJ4130/Natural Science Foundation of Hunan Province
- 81974532/National Natural Science Foundation of China
- 2022JJ10097/Science Fund for Distinguished Young Scholars of Hunan Province
- 21B0014/Scientific Research Foundation of Hunan Provincial Education Department
LinkOut - more resources
Full Text Sources

