Clinical Variant Reclassification in Hereditary Disease Genetic Testing
- PMID: 39504018
- PMCID: PMC11541632
- DOI: 10.1001/jamanetworkopen.2024.44526
Clinical Variant Reclassification in Hereditary Disease Genetic Testing
Abstract
Importance: Because accurate and consistent classification of DNA sequence variants is fundamental to germline genetic testing, understanding patterns of initial variant classification (VC) and subsequent reclassification from large-scale, empirical data can help improve VC methods, promote equity among race, ethnicity, and ancestry (REA) groups, and provide insights to inform clinical practice.
Objectives: To measure the degree to which initial VCs met certainty thresholds set by professional guidelines and quantify the rates of, the factors associated with, and the impact of reclassification among more than 2 million variants.
Design, setting, and participants: This cohort study used clinical multigene panel and exome sequencing data from diagnostic testing for hereditary disorders, carrier screening, or preventive genetic screening from individuals for whom genetic testing was performed between January 1, 2015, and June 30, 2023.
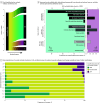
Exposure: DNA variants were classified into 1 of 5 categories: benign, likely benign, variant of uncertain significance (VUS), likely pathogenic, or pathogenic.
Main outcomes and measures: The main outcomes were accuracy of classifications, rates and directions of reclassifications, evidence contributing to reclassifications, and their impact across different clinical areas and REA groups. One-way analysis of variance followed by post hoc pairwise Tukey honest significant difference tests were used to analyze differences among means, and pairwise Pearson χ2 tests with Bonferroni corrections were used to compare categorical variables among groups.
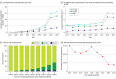
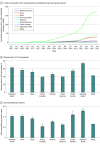
Results: The cohort comprised 3 272 035 individuals (median [range] age, 44 [0-89] years; 2 240 506 female [68.47%] and 1 030 729 male [31.50%]; 216 752 Black [6.62%]; 336 414 Hispanic [10.28%]; 1 804 273 White [55.14%]). Among 2 051 736 variants observed over 8 years in this cohort, 94 453 (4.60%) were reclassified. Some variants were reclassified more than once, resulting in 105 172 total reclassification events. The majority (64 752 events [61.65%]) were changes from VUS to either likely benign, benign, likely pathogenic, or pathogenic categories. An additional 37.66% of reclassifications (39 608 events) were gains in classification certainty to terminal categories (ie, likely benign to benign and likely pathogenic to pathogenic). Only a small fraction (663 events [0.63%]) moved toward less certainty, or very rarely (61 events [0.06%]) were classification reversals. When normalized by the number of individuals tested, VUS reclassification rates were higher among specific underrepresented REA populations (Ashkenazi Jewish, Asian, Black, Hispanic, Pacific Islander, and Sephardic Jewish). Approximately one-half of VUS reclassifications (37 074 of 64 840 events [57.18%]) resulted from improved use of data from computational modeling.
Conclusions and relevance: In this cohort study of individuals undergoing genetic testing, the empirically estimated accuracy of pathogenic, likely pathogenic, benign, and likely benign classifications exceeded the certainty thresholds set by current VC guidelines, suggesting the need to reevaluate definitions of these classifications. The relative contribution of various strategies to resolve VUS, including emerging machine learning-based computational methods, RNA analysis, and cascade family testing, provides useful insights that can be applied toward further improving VC methods, reducing the rate of VUS, and generating more definitive results for patients.
Conflict of interest statement
Figures
References
-
- Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 - DOI - PMC - PubMed
-
- Clinical Genome Resource . Overview of DRAFT ACMG/AMP v4 sequence variant guidelines. ClinGen. Accessed March 26, 2024. https://clinicalgenome.org/tools/clingen-summer-workshop-series-2023/sep...
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

