Identification of small molecule agonists of fetal hemoglobin expression for the treatment of sickle cell disease
- PMID: 39504332
- PMCID: PMC11540224
- DOI: 10.1371/journal.pone.0307049
Identification of small molecule agonists of fetal hemoglobin expression for the treatment of sickle cell disease
Abstract
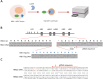
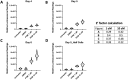
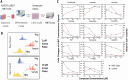
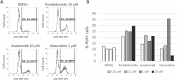
Induction of fetal hemoglobin (HbF) has been shown to be a viable therapeutic approach to treating sickle cell disease and potentially other β-hemoglobinopathies. To identify targets and target-modulating small molecules that enhance HbF expression, we engineered a human umbilical-derived erythroid progenitor reporter cell line (HUDEP2_HBG1_HiBiT) by genetically tagging a HiBiT peptide to the carboxyl (C)-terminus of the endogenous HBG1 gene locus, which codes for γ-globin protein, a component of HbF. Employing this reporter cell line, we performed a chemogenomic screen of approximately 5000 compounds annotated with known targets or mechanisms that have achieved clinical stage or approval by the US Food and Drug Administration (FDA). Among them, 10 compounds were confirmed for their ability to induce HbF in the HUDEP2 cell line. These include several known HbF inducers, such as pomalidomide, lenalidomide, decitabine, idoxuridine, and azacytidine, which validate the translational nature of this screening platform. We identified avadomide, autophinib, triciribine, and R574 as novel HbF inducers from these screens. We orthogonally confirmed HbF induction activities of the top hits in both parental HUDEP2 cells as well as in human primary CD34+ hematopoietic stem and progenitor cells (HSPCs). Further, we demonstrated that pomalidomide and avadomide, but not idoxuridine, induced HbF expression through downregulation of several transcriptional repressors such as BCL11A, ZBTB7A, and IKZF1. These studies demonstrate a robust phenotypic screening workflow that can be applied to large-scale small molecule profiling campaigns for the discovery of targets and pathways, as well as novel therapeutics for sickle cell disease and other β-hemoglobinopathies.
Copyright: © 2024 Yang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
Takeda Development Center Americas, Inc. provided sponsorship and financial support for this study. All the authors are employees of Takeda Pharmaceutical Industries, Ltd., and had equity ownership with Takeda Pharmaceutical Industries, Ltd. The Takeda commercial affiliation does not alter our adherence to PLOS ONE policies on sharing data and materials. There are no patents, products in development or marketed products associated with this research to declare.
Figures

References
-
- Longo D.L., et al.., Sickle cell disease. N Engl J Med, 2017. 20(April (376 (16))): p. 1561–1573. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

