Dormancy-inducing 3D engineered matrix uncovers mechanosensitive and drug-protective FHL2-p21 signaling axis
- PMID: 39504377
- PMCID: PMC11540038
- DOI: 10.1126/sciadv.adr3997
Dormancy-inducing 3D engineered matrix uncovers mechanosensitive and drug-protective FHL2-p21 signaling axis
Abstract
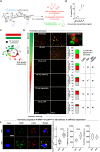
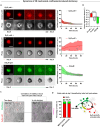
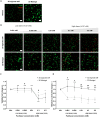
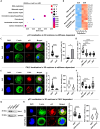
Solid cancers frequently relapse with distant metastasis, despite local and systemic treatment. Cellular dormancy has been identified as an important mechanism underlying drug resistance enabling late relapse. Therefore, relapse from invisible, minimal residual cancer of seemingly disease-free patients call for in vitro models of dormant cells suited for drug discovery. Here, we explore dormancy-inducing 3D engineered matrices, which generate mechanical confinement and induce growth arrest and survival against chemotherapy in cancer cells. We characterized the dormant phenotype of solitary cells by P-ERKlow:P-p38high dormancy signaling ratio, along with Ki67- expression. As underlying mechanism, we identified stiffness-dependent nuclear localization of the four-and-a-half LIM domain 2 (FHL2) protein, leading to p53-independent high p21Cip1/Waf1 nuclear expression, validated in murine and human tissue. Suggestive of a resistance-causing role, cells in the dormancy-inducing matrix became sensitive against chemotherapy upon FHL2 down-regulation. Thus, our biomaterial-based approach will enable systematic screens for previously unidentified compounds suited to eradicate potentially relapsing dormant cancer cells.
Figures



References
-
- Klein C. A., Cancer progression and the invisible phase of metastatic colonization. Nat. Rev. Cancer 20, 681–694 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

