Generalizable and replicable brain-based predictions of cognitive functioning across common psychiatric illness
- PMID: 39504381
- PMCID: PMC11540040
- DOI: 10.1126/sciadv.adn1862
Generalizable and replicable brain-based predictions of cognitive functioning across common psychiatric illness
Abstract
A primary aim of computational psychiatry is to establish predictive models linking individual differences in brain functioning with symptoms. In particular, cognitive impairments are transdiagnostic, treatment resistant, and associated with poor outcomes. Recent work suggests that thousands of participants may be necessary for the accurate and reliable prediction of cognition, questioning the utility of most patient collection efforts. Here, using a transfer learning framework, we train a model on functional neuroimaging data from the UK Biobank to predict cognitive functioning in three transdiagnostic samples (ns = 101 to 224). We demonstrate prediction performance in all three samples comparable to that reported in larger prediction studies and a boost of up to 116% relative to classical models trained directly in the smaller samples. Critically, the model generalizes across datasets, maintaining performance when trained and tested across independent samples. This work establishes that predictive models derived in large population-level datasets can boost the prediction of cognition across clinical studies.
Figures
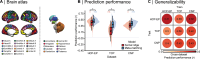
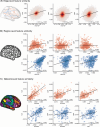

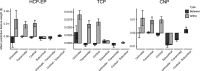
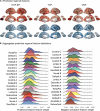
References
-
- Varoquaux G., Cross-validation failure: Small sample sizes lead to large error bars. Neuroimage 180, 68–77 (2018). - PubMed
-
- Whelan R., Garavan H., When optimism hurts: Inflated predictions in psychiatric neuroimaging. Biol. Psychiatry 75, 746–748 (2014). - PubMed
-
- Abramovitch A., Short T., Schweiger A., The C Factor: Cognitive dysfunction as a transdiagnostic dimension in psychopathology. Clin. Psychol. Rev. 86, 102007 (2021). - PubMed
-
- East-Richard C., Mercier A. R., Nadeau D., Cellard C., Transdiagnostic neurocognitive deficits in psychiatry: A review of meta-analyses. Can. Psychol. 61, 190–214 (2020).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

