One-ended and two-ended breaks at nickase-broken replication forks
- PMID: 39504607
- PMCID: PMC11681922
- DOI: 10.1016/j.dnarep.2024.103783
One-ended and two-ended breaks at nickase-broken replication forks
Abstract
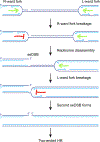
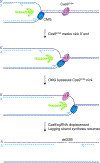
Replisome collision with a nicked parental DNA template can lead to the formation of a replication-associated double strand break (DSB). How this break is repaired has implications for cancer initiation, cancer therapy and therapeutic gene editing. Recent work shows that collision of a replisome with a nicked DNA template can give rise to either a single-ended (se) or a double-ended (de)DSB, with potentially divergent effects on repair pathway choice and genomic instability. Emerging evidence suggests that the biochemical environment of the broken mammalian replication fork may be specialized in such a way as to skew repair in favor of homologous recombination at the expense of non-homologous end joining.
Keywords: BRCA1; Break-induced replication; Homologous recombination; Mammalian DSB repair; One-ended break; Replication fork breakage.
Copyright © 2024 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

