3D chromatin maps of a brown alga reveal U/V sex chromosome spatial organization
- PMID: 39505852
- PMCID: PMC11541908
- DOI: 10.1038/s41467-024-53453-5
3D chromatin maps of a brown alga reveal U/V sex chromosome spatial organization
Abstract
Nuclear three dimensional (3D) folding of chromatin structure has been linked to gene expression regulation and correct developmental programs, but little is known about the 3D architecture of sex chromosomes within the nucleus, and how that impacts their role in sex determination. Here, we determine the sex-specific 3D organization of the model brown alga Ectocarpus chromosomes at 2 kb resolution, by mapping long-range chromosomal interactions using Hi-C coupled with Oxford Nanopore long reads. We report that Ectocarpus interphase chromatin exhibits a non-Rabl conformation, with strong contacts among telomeres and among centromeres, which feature centromere-specific LTR retrotransposons. The Ectocarpus chromosomes do not contain large local interactive domains that resemble TADs described in animals, but their 3D genome organization is largely shaped by post-translational modifications of histone proteins. We show that the sex determining region (SDR) within the U and V chromosomes are insulated and span the centromeres and we link sex-specific chromatin dynamics and gene expression levels to the 3D chromatin structure of the U and V chromosomes. Finally, we uncover the unique conformation of a large genomic region on chromosome 6 harboring an endogenous viral element, providing insights regarding the impact of a latent giant dsDNA virus on the host genome's 3D chromosomal folding.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
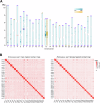
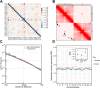

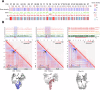
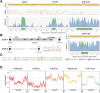
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 864038/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- GBMF11489/Gordon and Betty Moore Foundation (Gordon E. and Betty I. Moore Foundation)
- 101109906/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources
Miscellaneous

