Fine-mapping and molecular characterisation of primary sclerosing cholangitis genetic risk loci
- PMID: 39505854
- PMCID: PMC11541731
- DOI: 10.1038/s41467-024-53602-w
Fine-mapping and molecular characterisation of primary sclerosing cholangitis genetic risk loci
Abstract
Genome-wide association studies of primary sclerosing cholangitis have identified 23 susceptibility loci. The majority of these loci reside in non-coding regions of the genome and are thought to exert their effect by perturbing the regulation of nearby genes. Here, we aim to identify these genes to improve the biological understanding of primary sclerosing cholangitis, and nominate potential drug targets. We first build an eQTL map for six primary sclerosing cholangitis-relevant T-cell subsets obtained from the peripheral blood of primary sclerosing cholangitis and ulcerative colitis patients. These maps identify 10,459 unique eGenes, 87% of which are shared across all six primary sclerosing cholangitis T-cell types. We then search for colocalisations between primary sclerosing cholangitis loci and eQTLs and undertake Bayesian fine-mapping to identify disease-causing variants. In this work, colocalisation analyses nominate likely primary sclerosing cholangitis effector genes and biological mechanisms at five non-coding (UBASH3A, PRKD2, ETS2 and AP003774.1/CCDC88B) and one coding (SH2B3) primary sclerosing cholangitis loci. Through fine-mapping we identify likely causal variants for a third of all primary sclerosing cholangitis-associated loci, including two to single variant resolution.
© 2024. The Author(s).
Conflict of interest statement
C.A.A. has received consultancy fees from Genomics plc and BrideBio Ltd. All other authors declare no competing interests.
Figures

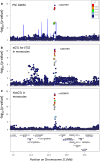

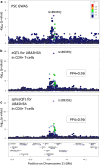

References
-
- Claessen, M. M. et al. High lifetime risk of cancer in primary sclerosing cholangitis. J. Hepatol.50, 158–164 (2009). - PubMed
-
- Sano, H. et al. Clinical characteristics of inflammatory bowel disease associated with primary sclerosing cholangitis. J. Hepatobiliary Pancreat. Sci.18, 154–161 (2011). - PubMed
-
- Boonstra, K. et al. Primary sclerosing cholangitis is associated with a distinct phenotype of inflammatory bowel disease. Inflamm. Bowel Dis.18, 2270–2276 (2012). - PubMed
-
- Boonstra, K., Beuers, U. & Ponsioen, C. Y. Epidemiology of primary sclerosing cholangitis and primary biliary cirrhosis: a systematic review. J. Hepatol.56, 1181–1188 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

