AMFR-mediated Flavivirus NS2A ubiquitination subverts ER-phagy to augment viral pathogenicity
- PMID: 39505910
- PMCID: PMC11541587
- DOI: 10.1038/s41467-024-54010-w
AMFR-mediated Flavivirus NS2A ubiquitination subverts ER-phagy to augment viral pathogenicity
Abstract
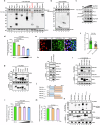
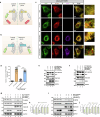
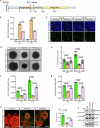
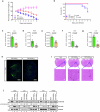
Flaviviruses strategically utilize the endoplasmic reticulum (ER) in their replication cycles. However, the role of ER autophagy (ER-phagy) in viral replication process remains poorly understood. Here, we reveal that prolonged Zika virus (ZIKV) infection results from the degradation of ER-phagy receptor FAM134B, facilitated by viral NS2A protein. Mechanistically, ER-localized NS2A undergoes K48-linked polyubiquitination at lysine (K) 56 by E3 ligase AMFR. Ubiquitinated NS2A binds to FAM134B and AMFR orchestrates the degradation of NS2A-FAM134B complexes. AMFR-catalyzed NS2A ubiquitination not only targets FAM134B degradation but also hinders the FAM134B-AMFR axis. Notably, a recombinant ZIKV mutant (ZIKV-NS2AK56R), lacking ubiquitination and ER-phagy inhibition, exhibits attenuation in ZIKV-induced microcephalic phenotypes in human brain organoids and replicates less efficiently, resulting in weakened pathogenesis in mouse models. In this work, our mechanistic insights propose that flaviviruses manipulate ER-phagy to modulate ER turnover, driving viral infection. Furthermore, AMFR-mediated flavivirus NS2A ubiquitination emerges as a potential determinant of viral pathogenecity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

