A metagenomic catalogue of the ruminant gut archaeome
- PMID: 39505912
- PMCID: PMC11542040
- DOI: 10.1038/s41467-024-54025-3
A metagenomic catalogue of the ruminant gut archaeome
Erratum in
-
Publisher Correction: A metagenomic catalogue of the ruminant gut archaeome.Nat Commun. 2025 Jan 6;16(1):408. doi: 10.1038/s41467-024-55731-8. Nat Commun. 2025. PMID: 39762210 Free PMC article. No abstract available.
Abstract
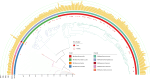
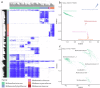
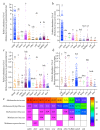
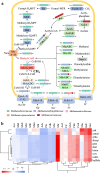
While the ruminant gut archaeome regulates the gut microbiota and hydrogen balance, it is also a major producer of the greenhouse gas methane. However, ruminant gut archaeome diversity within the gastrointestinal tract (GIT) of ruminant animals worldwide remains largely underexplored. Here, we construct a catalogue of 998 unique archaeal genomes recovered from the GITs of ruminants, utilizing 2270 metagenomic samples across 10 different ruminant species. Most of the archaeal genomes (669/998 = 67.03%) belong to Methanobacteriaceae and Methanomethylophilaceae (198/998 = 19.84%). We recover 47/279 previously undescribed archaeal genomes at the strain level with completeness of >80% and contamination of <5%. We also investigate the archaeal gut biogeography across various ruminants and demonstrate that archaeal compositional similarities vary significantly by breed and gut location. The catalogue contains 42,691 protein clusters, and the clustering and methanogenic pathway analysis reveal strain- and host-specific dependencies among ruminant animals. We also find that archaea potentially carry antibiotic and metal resistance genes, mobile genetic elements, virulence factors, quorum sensors, and complex archaeal viromes. Overall, this catalogue is a substantial repository for ruminant archaeal recourses, providing potential for advancing our understanding of archaeal ecology and discovering strategies to regulate methane production in ruminants.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- IPCC. Sections. In: Climate Change 2023: Synthesis Report. Contribution of Working Groups I, II and III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change [Core Writing Team, H. Lee and J. Romero (eds.)]. IPCC, Geneva, Switzerland, 35-115 (2023).
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

