Public health concern of antimicrobial resistance and virulence determinants in E. coli isolates from oysters in Egypt
- PMID: 39505944
- PMCID: PMC11541584
- DOI: 10.1038/s41598-024-77519-y
Public health concern of antimicrobial resistance and virulence determinants in E. coli isolates from oysters in Egypt
Abstract
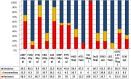
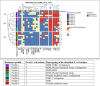
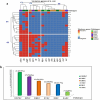
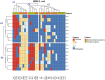
The emergence of critical-priority E. coli, carrying a wide array of resistance and virulence factors through food sources, poses a significant challenge to public health. This study aimed to investigate the potential role of oysters sold in Egypt as a source for E. coli, identify their resistance and virulence-associated gene profiles, and assess associated zoonotic risks. A total of 33 pooled fresh oyster samples were obtained from various retail fish markets in Egypt and examined bacteriologically for the presence of E. coli. Antimicrobial resistance was performed by the disk-diffusion method, and the multiple antibiotic resistance index (MAR) was calculated. All isolates were screened for extended-spectrum beta-lactamase (ESBL) (blaTEM, blaSHV, blaCTX-M, and blaOXA-1), plasmid-mediated AmpC blaCMY-2, and carbapenemases (blaKPC, blaNDM, blaVIM, and blaOXA-48) genes by Polymerase chain reaction. Moreover, the presence of virulence-encoding genes was investigated. The virulent MDR strains were clustered using R with the pheatmap package. The prevalence of E. coli was 72.7% (24 out of 33), with 66.7% of the isolates classified as multi-drug resistant, and 75% exhibited MAR values exceeding the 0.2 threshold. Different antimicrobial sensitivity phenotypes and genotype profiles were identified in E. coli isolates. The most prevalent gene detected among all isolates was blaTEM (22/24, 91.7%). Notably, all non-ESBL producers were positive for blaCMY2. Carbapenem-resistant and carbapenem-intermediate strains were carbapenemase producers, with the predominance of the blaKPC gene (11/24, 45.8%). Remarkably, twelve out of sixteen virulence genes were identified, with papC (21/24, 87.5%) and sfa (16/24, 66.7%) genes being the most prevalent. Most isolates carry virulence genes primarily associated with extra-intestinal pathogenic E. coli (ExPEC) (87.5%) and enteropathogenic (EPEC) (70.8%) pathotypes. Four E. coli isolates exhibit cluster patterns. This study provides the first insight into the emergence of virulent MDR E. coli among oysters in Egypt. It underscores the potential role of oysters as a source for disseminating these strains within aquatic ecosystems, presenting a possible threat to public health.
Keywords: E. Coli pathotypes; Carbapenems; Extended-spectrum β-lactamases; Multi-drug resistance; Plasmid AmpC; Virulence factors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

