Specific human gene expression in response to infection is an effective marker for diagnosis of latent and active tuberculosis
- PMID: 39505948
- PMCID: PMC11541504
- DOI: 10.1038/s41598-024-77164-5
Specific human gene expression in response to infection is an effective marker for diagnosis of latent and active tuberculosis
Abstract
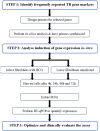
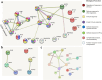
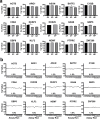
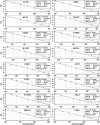
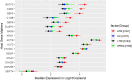
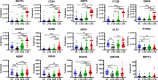
RNA sequencing and microarray analysis revealed transcriptional markers expressed in whole blood can differentiate active pulmonary TB (ATB) from other respiratory diseases (ORDs), and latent TB infection (LTBI) from healthy controls (HC). Here we describe a streamlined reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) assay that could be applied at near point-of-care for diagnosing and distinguishing ATB from ORDs and LTBI from HC. A literature review was undertaken to identify the most plausible host-gene markers (HGM) of TB infection. Primers, and dual labelled hydrolysis probes were designed and analytically evaluated for accuracy in an in-vitro model of infection using a lung fibroblast cell-line. Best performing genes were multiplexed into panels of 2-4 targets and taken forward for clinical evaluation. Mycobacteria Growth Indicator Tube and QuantiFERON-TB Gold Plus were used as reference tests for ATB and LTBI respectively. A total of 16 HGM were selected and incorporated into five multiplex RT-qPCR panels. PCR assay efficiency of all evaluated targets was ≥ 90% with a median analytical sensitivity of 292 copies/µl [IQR: 215.0-358.3 copies/µl], and a median limit of quantification of 61.7 copies/µl [IQR: 29.4-176.3 copies/µl]. Clinically, ATB was characterised by higher gene expression than ORDs, while LTBI was associated with lower gene expression than HC, Kruskal-Wallis p < 0.0001. Crucially, BATF2, CD64, GBP5, C1QB, GBP6, DUSP3, and GAS6 exhibited high differentiative ability for ATB from ORDs, LTBI or HC while KLF2, PTPRC, NEMF, ASUN, and ZNF296 independently discriminated LTBI from HC. Our results show that different HGM maybe required for ATB and LTBI differentiation from ORDs or HC respectively and demonstrate the feasibility of host gene-based RT-qPCR to diagnose ATB and LTBI at near point-of-care.
Keywords: Active tuberculosis; Diagnosis; Host gene expression; Latent tuberculosis; Reverse transcriptase-quantitative PCR.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- WHO. WHO. Early detection of Tuberculosis: An overview of approaches, guidelines and tools. WHO/HTM/STB/PSI/2011.21. 1–32 at. (2011).
-
- WHO. High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting. WHO Meeting Report (2014). https://iris.who.int/handle/10665/135617
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

