Cellular ATP demand creates metabolically distinct subpopulations of mitochondria
- PMID: 39506109
- PMCID: PMC11869630
- DOI: 10.1038/s41586-024-08146-w
Cellular ATP demand creates metabolically distinct subpopulations of mitochondria
Abstract
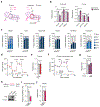
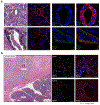
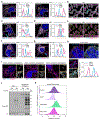
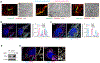
Mitochondria serve a crucial role in cell growth and proliferation by supporting both ATP synthesis and the production of macromolecular precursors. Whereas oxidative phosphorylation (OXPHOS) depends mainly on the oxidation of intermediates from the tricarboxylic acid cycle, the mitochondrial production of proline and ornithine relies on reductive synthesis1. How these competing metabolic pathways take place in the same organelle is not clear. Here we show that when cellular dependence on OXPHOS increases, pyrroline-5-carboxylate synthase (P5CS)-the rate-limiting enzyme in the reductive synthesis of proline and ornithine-becomes sequestered in a subset of mitochondria that lack cristae and ATP synthase. This sequestration is driven by both the intrinsic ability of P5CS to form filaments and the mitochondrial fusion and fission cycle. Disruption of mitochondrial dynamics, by impeding mitofusin-mediated fusion or dynamin-like-protein-1-mediated fission, impairs the separation of P5CS-containing mitochondria from mitochondria that are enriched in cristae and ATP synthase. Failure to segregate these metabolic pathways through mitochondrial fusion and fission results in cells either sacrificing the capacity for OXPHOS while sustaining the reductive synthesis of proline, or foregoing proline synthesis while preserving adaptive OXPHOS. These findings provide evidence of the key role of mitochondrial fission and fusion in maintaining both oxidative and reductive biosyntheses in response to changing nutrient availability and bioenergetic demand.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: C.M.R. has consulted regarding oncology drug development with Amgen, AstraZeneca, Chugai, D2G, Daiichi Sankyo, Hoffman-La Roche, Jazz and Legend, and serves on the scientific advisory boards of Auron, Bridge Medicines, DISCO, Earli and Harpoon Therapeutics. C.B.T. is a founder of Agios Pharmaceuticals, and is on the board of directors of Regeneron and Charles River Laboratories. The remaining authors declare no competing interests.
Figures









References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

