Automated real-world data integration improves cancer outcome prediction
- PMID: 39506116
- PMCID: PMC11655358
- DOI: 10.1038/s41586-024-08167-5
Automated real-world data integration improves cancer outcome prediction
Abstract
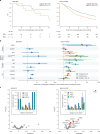
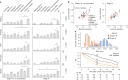
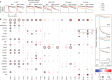
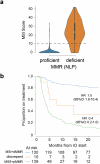
The digitization of health records and growing availability of tumour DNA sequencing provide an opportunity to study the determinants of cancer outcomes with unprecedented richness. Patient data are often stored in unstructured text and siloed datasets. Here we combine natural language processing annotations1,2 with structured medication, patient-reported demographic, tumour registry and tumour genomic data from 24,950 patients at Memorial Sloan Kettering Cancer Center to generate a clinicogenomic, harmonized oncologic real-world dataset (MSK-CHORD). MSK-CHORD includes data for non-small-cell lung (n = 7,809), breast (n = 5,368), colorectal (n = 5,543), prostate (n = 3,211) and pancreatic (n = 3,109) cancers and enables discovery of clinicogenomic relationships not apparent in smaller datasets. Leveraging MSK-CHORD to train machine learning models to predict overall survival, we find that models including features derived from natural language processing, such as sites of disease, outperform those based on genomic data or stage alone as tested by cross-validation and an external, multi-institution dataset. By annotating 705,241 radiology reports, MSK-CHORD also uncovers predictors of metastasis to specific organ sites, including a relationship between SETD2 mutation and lower metastatic potential in immunotherapy-treated lung adenocarcinoma corroborated in independent datasets. We demonstrate the feasibility of automated annotation from unstructured notes and its utility in predicting patient outcomes. The resulting data are provided as a public resource for real-world oncologic research.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: S.B.M. declares professional services and activities for Amgen, Clinical Care Options, Daiichi Sankyo, Elevation Oncology, MedPage Today, Novartis, Physicians’ Education Resource, Pinetree Therapeutics, Purple Biotech and Vindico Medical Education; and equity in McKesson. L.B. declares professional services and activities for the Cancer Prevention & Research Institute of Texas. M.R. declares professional services and activities (uncompensated) for Artios Pharma, AstraZeneca, Foundation Medicine, Pfizer and Tempus Labs; and professional services and activities for Change Healthcare, Clinical Education Alliance, Genome Quebec, MJH Associates and myMedEd. M.L. declares equity in and professional services and activities (uncompensated) for Paige.AI. D.B.S. declares professional services and activities for American Association for Cancer Research, BridgeBio, Fog Pharmaceuticals, Paige.AI, Pfizer, Rain Therapeutics; and equity in and professional services and activities for Elsie Biotechnologies, Fore Biotherapeutics, Function Oncology, Pyramid Biosciences and Scorpion Therapeutics. D.R.J. declares professional services and activities for AstraZeneca, Dava Oncology and MORE Health; and professional services and activities (uncompensated) for Merck & Co. D.G. declares professional services and activities for AstraZeneca, Grail, Johnson & Johnson, Med Learning Group, Medtronic and Varian Medical Systems. H.Y. declares professional services and activities for AbbVie, AstraZeneca, Black Diamond Therapeutics, Blueprint Medicines, C4 Therapeutics, Daiichi Sankyo, Ipsen Pharma, Janssen Pharmaceuticals, Taiho and Takeda Pharmaceuticals. R.Y. declares professional services and activities for Mirati Therapeutics and Zai Lab. W.A. declares professional services and activities for AstraZeneca, Clinical Education Alliance, Janssen Oncology and Touch Independent Medical Education. W.P. declares professional services and activities for Astellas. J.G.-A. declares professional services and activities for Ethicon; and equity in and professional services and activities for Intuitive Surgical. P.D.S. declares professional services and activities for the National Comprehensive Cancer Network and the National Institutes of Health. R. Levine declares equity, a fiduciary role or position and intellectual property rights in and professional services and activities (uncompensated) for Ajax Therapeutics; equity in Anovia Biosciences, Bakx Therapeutics, Epiphanes, Imago Biosciences and Syndax; professional services and activities for AstraZeneca, Genome Quebec, Goldman Sachs, Incyte, Janssen Pharmaceuticals and Jubilant Therapeutics; equity in and professional services and activities (uncompensated) for Auron Therapeutics and the Isoplexis Corporation; equity in and professional services and activities for C4 Therapeutics, Kurome Therapeutics, Mana Therapeutics, Mission Bio, Prelude Therapeutics, Scorpion Therapeutics, Zentalis Pharmaceuticals; intellectual property rights in the Cure Breast Cancer Foundation and Epizyme; professional services and activities (uncompensated) for the ECOG-ACRIN Cancer Research Group; equity and a fiduciary role or position in and professional services and activities (uncompensated) for Qiagen; and a fiduciary role or position in and professional services and activities for The Mark Foundation. C.M.R. declares professional services and activities for Amgen, AstraZeneca, Bridge Medicines, D2G Oncology, Harpoon Therapeutics and Jazz Pharmaceuticals; intellectual property rights in Daiichi Sankyo; and equity in Earli. M.F.B. declares professional services and activities for AstraZeneca and Paige.AI; professional services and activities (uncompensated) for JCO Precision Oncology and the Journal of Molecular Diagnostics; and intellectual property rights in SOPHiA GENETICS. P.R. declares professional services and activities for Biovica, Inivata, Novartis, Prelude Therapeutics and SAGA Diagnostics; professional services and activities (uncompensated) for Guardant Health, Paige.AI and Tempus Labs; and equity, a fiduciary role or position and intellectual property rights in Odyssey Biosciences. B.T.L. declares professional services and activities (uncompensated) for Amgen, the Asia Society, AstraZeneca, Bolt Biotherapeutics and Daiichi Sankyo; and intellectual property rights in Karger Publishers and Shanghai Jiao Tong University Press. G.J.R. declares professional services and activities (uncompensated) for the American Association for Cancer Research, the American Society of Clinical Oncology, Mirati Therapeutics, Pfizer, Takeda Pharmaceuticals and Verastem; and professional services and activities for Harborside Press, MJH Associates, the National Comprehensive Cancer Network, Phillips Gilmore Oncology Communications, Research to Practice and Triptych Health Partners. H.S. declares professional services and activities for Bayer, Pfizer, Regeneron Pharmaceuticals, Sanofi and WCG Oncology; and intellectual property rights in Elucida Oncology. J.S.R.-F. is an employee of AstraZeneca, has served as a consultant for Goldman Sachs, Paige.AI and REPARE Therapeutics; and has served as an adviser for Roche, Genentech, Roche Tissue Diagnostics, Ventana, Novartis, InVicro, GRAIL, Goldman Sachs, Paige.AI and Volition RX. J. Gao and M.C. are employees of Caris.
Figures






References
-
- Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst.30, 6000–6010 (2017)
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

