Diverse Genome Structures among Eukaryotes May Have Arisen in Response to Genetic Conflict
- PMID: 39506510
- PMCID: PMC11606643
- DOI: 10.1093/gbe/evae239
Diverse Genome Structures among Eukaryotes May Have Arisen in Response to Genetic Conflict
Abstract
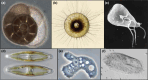
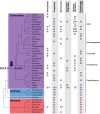
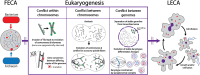
In contrast to the typified view of genome cycling only between haploidy and diploidy, there is evidence from across the tree of life of genome dynamics that alter both copy number (i.e. ploidy) and chromosome complements. Here, we highlight examples of such processes, including endoreplication, aneuploidy, inheritance of extrachromosomal DNA, and chromatin extrusion. Synthesizing data on eukaryotic genome dynamics in diverse extant lineages suggests the possibility that such processes were present before the last eukaryotic common ancestor. While present in some prokaryotes, these features appear exaggerated in eukaryotes where they are regulated by eukaryote-specific innovations including the nucleus, complex cytoskeleton, and synaptonemal complex. Based on these observations, we propose a model by which genome conflict drove the transformation of genomes during eukaryogenesis: from the origin of eukaryotes (i.e. first eukaryotic common ancestor) through the evolution of last eukaryotic common ancestor.
Keywords: aneuploidy; eukaryogenesis; genetic conflict; genome dynamics; meiosis; polyploidy.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
-
- Aravind L, Anantharaman V, Zhang D, De Souza R, Iyer L. Gene flow and biological conflict systems in the origin and evolution of eukaryotes. Front Cell Infect Microbiol. 2012:2:89. https://www.frontiersin.org/articles/10.3389/fcimb.2012.00089 10.3389/fcimb.2012.00089. - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

