Characterization of Single-Cell Cis-regulatory Elements Informs Implications for Cell Differentiation
- PMID: 39506564
- PMCID: PMC11580522
- DOI: 10.1093/gbe/evae241
Characterization of Single-Cell Cis-regulatory Elements Informs Implications for Cell Differentiation
Abstract
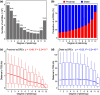
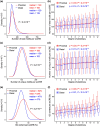
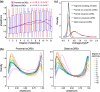
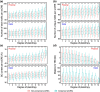
Cis-regulatory elements govern the specific patterns and dynamics of gene expression in cells during development, which are the fundamental mechanisms behind cell differentiation. However, the genomic characteristics of single-cell cis-regulatory elements closely linked to cell differentiation during development remain unclear. To explore this, we systematically analyzed ∼250,000 putative single-cell cis-regulatory elements obtained from snATAC-seq analysis of the developing mouse cerebellum. We found that over 80% of these single-cell cis-regulatory elements show pleiotropic effects, being active in 2 or more cell types. The pleiotropic degrees of proximal and distal single-cell cis-regulatory elements are positively correlated with the density and diversity of transcription factor binding motifs and GC content. There is a negative correlation between the pleiotropic degrees of single-cell cis-regulatory elements and their distances to the nearest transcription start sites, and proximal single-cell cis-regulatory elements display higher relevance strengths than distal ones. Furthermore, both proximal and distal single-cell cis-regulatory elements related to cell differentiation exhibit enhanced sequence-level evolutionary conservation, increased density and diversity of transcription factor binding motifs, elevated GC content, and greater distances from their nearest genes. Together, our findings reveal the general genomic characteristics of putative single-cell cis-regulatory elements and provide insights into the genomic and evolutionary mechanisms by which single-cell cis-regulatory elements regulate cell differentiation during development.
Keywords: CRE; cell differentiation; genomic characteristics; pleiotropy.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest The authors declare no competing interests.
Figures
Similar articles
-
Definition of regulatory elements and transcription factors controlling porcine immune cell gene expression at single cell resolution using single nucleus ATAC-seq.Genomics. 2024 Nov;116(6):110944. doi: 10.1016/j.ygeno.2024.110944. Epub 2024 Sep 24. Genomics. 2024. PMID: 39326643
-
Synthetic and genomic regulatory elements reveal aspects of cis-regulatory grammar in mouse embryonic stem cells.Elife. 2020 Feb 11;9:e41279. doi: 10.7554/eLife.41279. Elife. 2020. PMID: 32043966 Free PMC article.
-
Profiling of chromatin accessibility and identification of general cis-regulatory mechanisms that control two ocular lens differentiation pathways.Epigenetics Chromatin. 2019 May 3;12(1):27. doi: 10.1186/s13072-019-0272-y. Epigenetics Chromatin. 2019. PMID: 31053165 Free PMC article.
-
Single-cell analysis of cis-regulatory elements.Curr Opin Plant Biol. 2022 Feb;65:102094. doi: 10.1016/j.pbi.2021.102094. Epub 2021 Aug 12. Curr Opin Plant Biol. 2022. PMID: 34390932 Review.
-
The Hematopoietic Stem and Progenitor Cell Cistrome: GATA Factor-Dependent cis-Regulatory Mechanisms.Curr Top Dev Biol. 2016;118:45-76. doi: 10.1016/bs.ctdb.2016.01.002. Epub 2016 Feb 26. Curr Top Dev Biol. 2016. PMID: 27137654 Free PMC article. Review.
Cited by
-
Dysregulated Gene Expression: A Candidate Mechanism for Anxiety Disorders.J Psychiatr Brain Sci. 2025;10(3):e250004. doi: 10.20900/jpbs.20250004. Epub 2025 Jun 25. J Psychiatr Brain Sci. 2025. PMID: 40689083 Free PMC article.
References
-
- Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva Pérez N, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022:50(D1):D165–D173. 10.1093/nar/gkab1113. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

