Single cell RNA-seq reveals cellular and transcriptional heterogeneity in the splenic CD11b+Ly6Chigh monocyte population expanded in sepsis-surviving mice
- PMID: 39506629
- PMCID: PMC11539566
- DOI: 10.1186/s10020-024-00970-0
Single cell RNA-seq reveals cellular and transcriptional heterogeneity in the splenic CD11b+Ly6Chigh monocyte population expanded in sepsis-surviving mice
Abstract
Background: Sepsis survivors exhibit immune dysregulation that contributes to poor long-term outcomes. Phenotypic and functional alterations within the myeloid compartment are believed to be a contributing factor. Here we dissect the cellular and transcriptional heterogeneity of splenic CD11b+Ly6Chigh myeloid cells that are expanded in mice that survive the cecal ligation and puncture (CLP) murine model of polymicrobial sepsis to better understand the basis of immune dysregulation in sepsis survivors.
Methods: Sham or CLP surgeries were performed on C57BL/6J and BALB/c mice. Four weeks later splenic CD11b+Ly6Chigh cells from both groups were isolated for phenotypic (flow cytometry) and functional (phagocytosis and glycolysis) characterization and RNA was obtained for single-cell RNA-seq (scRNA-seq) and subsequent analysis.
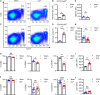
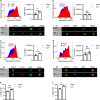
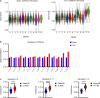
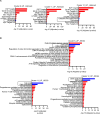
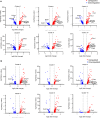
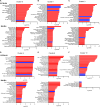
Results: CD11b+Ly6Chigh cells from sham and CLP surviving mice exhibit phenotypic and functional differences that relate to immune function, some of which are observed in both C57BL/6J and BALB/c strains and others that are not. To dissect disease-specific and strain-specific distinctions within the myeloid compartment, scRNA-seq analysis was performed on CD11b+Ly6Chigh cells from C57BL/6J and BALB/c sham and CLP mice. Uniform Manifold Approximation and Projection from both strains identified 13 distinct clusters of sorted CD11b+Ly6Chigh cells demonstrating significant transcriptional heterogeneity and expressing gene signatures corresponding to classical-monocytes, non-classical monocytes, M1- or M2-like macrophages, dendritic-like cells, monocyte-derived dendritic-like cells, and proliferating monocytic myeloid-derived suppressor cells (M-MDSCs). Frequency plots showed that the percentages of proliferating M-MDSCs (clusters 8, 11 and 12) were increased in CLP mice compared to sham mice in both strains. Pathway and UCell score analysis in CLP mice revealed that cell cycle and glycolytic pathways were upregulated in proliferating M-MDSCs in both strains. Notably, granule protease genes were upregulated in M-MDSCs from CLP mice. ScRNA-seq analyses also showed that phagocytic pathways were upregulated in multiple clusters including the classical monocyte cluster, confirming the increased phagocytic capacity in CD11b+Ly6Chigh cells from CLP mice observed in ex vivo functional assays in C57BL/6J mice.
Conclusion: The splenic CD11b+Ly6Chigh myeloid populations expanded in survivors of CLP sepsis correspond to proliferating cells that have an increased metabolic demand and gene signatures consistent with M-MDSCs, a population known to have immunosuppressive capacity.
Keywords: CD11b+Ly6Chigh; CLP; Glycolysis; Phagocytosis; Sepsis; scRNA-seq.
© 2024. The Author(s).
Conflict of interest statement
All authors are current or former staff members of The Feinstein Institutes for Medical Research. BD is on the board of Molecular Medicine and MR and MS are Associate Editors of Molecular Medicine. The authors declare that they have no other competing interests.
Figures


References
-
- Abbas AK, Lichtman AH, Pillai S, Baker DL, Baker A. Cellular and molecular immunology. 9th ed. Philadelphia: Elsevier; 2018. p. 325–48.
-
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–7. - PubMed
-
- Arts RJ, Joosten LA, Netea MG. Immunometabolic circuits in trained immunity. Semin Immunol. 2016;28(5):425–30. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

