DeepGR: a deep-learning prognostic model based on glycolytic radiomics for non-small cell lung cancer
- PMID: 39507025
- PMCID: PMC11535831
- DOI: 10.21037/tlcr-24-716
DeepGR: a deep-learning prognostic model based on glycolytic radiomics for non-small cell lung cancer
Abstract
Background: Glycolysis proved to have a prognostic value in lung cancer; however, to identify glycolysis-related genomic markers is expensive and challenging. This study aimed at identifying glycolysis-related computed tomography (CT) radiomics features to develop a deep-learning prognostic model for non-small cell lung cancer (NSCLC).
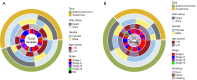
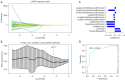
Methods: The study included 274 NSCLC patients from cohorts of The Second Affiliated Hospital of Soochow University (SZ; n=64), the Cancer Genome Atlas (TCGA)-NSCLC dataset (n=74), and the Gene Expression Omnibus dataset (n=136). Initially, the glycolysis enrichment scores were evaluated using a single-sample gene set enrichment analysis, and the cut-off values were optimized to investigate the prognostic potential of glycolysis genes. Radiomic features were then extracted using LIFEx software. The least absolute reduction and selection operator (LASSO) algorithm was employed to determine the glycolytic CT radiomics features. A deep-learning prognostic model was constructed by integrating CT radiomics and clinical features. The biological functions of the model were analyzed by incorporating RNA sequencing data.
Results: Kaplan-Meier curves indicated that elevated glycolysis levels were associated with poorer survival outcomes. The LASSO algorithm identified 11 radiomic features that were then selected for inclusion in the deep-learning model. They have shown significant discrimination capability in assessing glycolysis status, achieving an area under the curve value of 0.8442. The glycolysis-based radiomics deep-learning model was named the DeepGR model. This model was able to effectively predict the clinical outcomes of NSCLC patients with AUCs of 0.8760 and 0.8259 in the SZ and TCGA cohorts, respectively. High-risk DeepGR scores were strongly associated with poor overall survival, resting memory CD4+ T cells, and a high response to programmed cell death protein 1 immunotherapy.
Conclusions: The DeepGR model effectively predicted the prognosis of NSCLC patients.
Keywords: Non-small cell lung cancer (NSCLC); deep learning; glycolysis; prognostic model; radiomics.
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-716/coif). The authors have no conflicts of interest to declare.
Figures








References
LinkOut - more resources
Full Text Sources
Research Materials
