Role of CENPL, DARS2, and PAICS in determining the prognosis of patients with lung adenocarcinoma
- PMID: 39507047
- PMCID: PMC11535832
- DOI: 10.21037/tlcr-24-696
Role of CENPL, DARS2, and PAICS in determining the prognosis of patients with lung adenocarcinoma
Abstract
Background: Non-small cell lung cancer (NSCLC) accounts for about 85% of lung cancers, and is the leading cause of tumor-related death. Lung adenocarcinoma (LUAD) is the most prevalent subtype of NSCLC. Although significant progress of LUAD treatment has been made under multimodal strategies, the prognosis of advanced LUAD is still poor due to recurrence and metastasis. There is still a lack of reliable markers to evaluate the LUAD prognosis. This study aims to explore novel biomarkers and construct a prognostic model to predict the prognosis of LUAD patients.
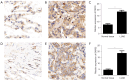
Methods: The Genomic Data Commons-The Cancer Genome Atlas-Lung Adenocarcinoma (GDC-TCGA-LUAD) dataset was downloaded from the University of California, Santa Cruz (UCSC) Xena browser. The GSE72094 and GSE13213 datasets and corresponding clinical information were downloaded from the Gene Expression Omnibus (GEO) database. By analyzing these datasets using DESeq2 R package and Limma R package, differentially expressed genes (DEGs) were found. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were used to analyze possible enrichment pathways. A protein-protein interaction (PPI) network was constructed to explore possible relationship among DEGs by using the STRING database. A survival analysis was performed to identify reliable prognostic genes using the Kaplan-Meier method. A multi-omics analysis was performed using the Gene Set Cancer Analysis (GSCA). The Tumor Immune Estimation Score (TIMER) database was used to analyze the association between prognostic genes and immune infiltration. A Spearman correlation analysis was conducted to examine the correlation between prognostic genes and drug sensitivity. A multivariate Cox regression was used to identify independent prognostic factors. Next, a nomogram was constructed using the rms R package. Finally, the expressions of aspartyl-tRNA synthetase 2 (DARS2) and phosphoribosyl aminoimidazole carboxylase (PAICS) were detected using immunohistochemistry (IHC).
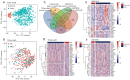
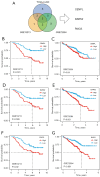
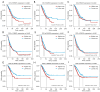
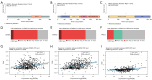
Results: We screened out 30 DEGs prior to functional enrichment and PPI network analysis revealing potential enrichment pathways and interactions of these DEGs. Then survival analysis revealed the CENPL, DARS2, and PAICS expression was negatively correlated with LUAD prognosis. Additionally, multi-omics analysis showed CENPL, DARS2, and PAICS expressions were significantly higher in LUAD tissues than normal tissues. CENPL, DARS2, and PAICS were all up-regulated in late stage and M1 stage. Correlation analysis indicated CENPL, DARS2, and PAICS may not be associated with activation or suppression of immune cells. Drug sensitivity analysis revealed many potentially effective drugs and small molecule compounds. Moreover, we successfully constructed a robust and stable nomogram by combining the DARS2 and PAICS expression with other clinicopathological variables. Finally, IHC results showed DARS2 and PAICS were significantly up-regulated in LUAD.
Conclusions: The CENPL, DARS2, and PAICS expression was negatively correlated with LUAD prognosis. A prognostic model, which integrated DARS2, PAICS, and other clinicopathological variables, was able to effectively predict LUAD patients prognosis.
Keywords: CENPL; DARS2; PAICS; lung adenocarcinoma (LUAD); prognosis.
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-696/coif). The authors have no conflicts of interest to declare.
Figures





References
LinkOut - more resources
Full Text Sources
Miscellaneous
