A multiorgan map of metabolic, signaling, and inflammatory pathways that coordinately control fasting glycemia in mice
- PMID: 39507247
- PMCID: PMC11539597
- DOI: 10.1016/j.isci.2024.111134
A multiorgan map of metabolic, signaling, and inflammatory pathways that coordinately control fasting glycemia in mice
Abstract
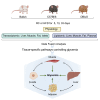
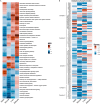
To identify the pathways that are coordinately regulated in pancreatic β cells, muscle, liver, and fat to control fasting glycemia we fed C57Bl/6, DBA/2, and Balb/c mice a regular chow or a high fat diet for 5, 13, and 33 days. Physiological, transcriptomic and lipidomic data were used in a data fusion approach to identify organ-specific pathways linked to fasting glycemia across all conditions investigated. In pancreatic islets, constant insulinemia despite higher glycemic levels was associated with reduced expression of hormone and neurotransmitter receptors, OXPHOS, cadherins, integrins, and gap junction mRNAs. Higher glycemia and insulin resistance were associated, in muscle, with decreased insulin signaling, glycolytic, Krebs' cycle, OXPHOS, and endo/exocytosis mRNAs; in hepatocytes, with reduced insulin signaling, branched chain amino acid catabolism and OXPHOS mRNAs; in adipose tissue, with increased innate immunity and lipid catabolism mRNAs. These data provide a resource for further studies of interorgan communication in glucose homeostasis.
Keywords: Bioinformatics; Omics; Physiology; Transcriptomics.
© 2024 The Author(s).
Conflict of interest statement
M.G., C.K., and K.S. are employees of Lipotype GmbH.
Figures






References
LinkOut - more resources
Full Text Sources
Miscellaneous

