The dissemination of multidrug-resistant and hypervirulent Klebsiella pneumoniae clones across the Kingdom of Saudi Arabia
- PMID: 39508718
- PMCID: PMC11583321
- DOI: 10.1080/22221751.2024.2427793
The dissemination of multidrug-resistant and hypervirulent Klebsiella pneumoniae clones across the Kingdom of Saudi Arabia
Abstract
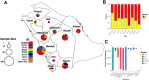
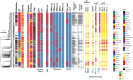
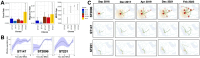
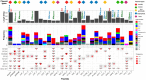
Klebsiella pneumoniae is a Gram-negative bacterium associated with a wide range of community- and hospital-acquired infections. The emergence of clonal hypervirulent strains resistant to last-resort antimicrobial agents has become a global concern. The Kingdom of Saudi Arabia (KSA), with its diverse population and high tourism traffic, serves as a platform where the spread of multidrug-resistant (MDR) strains are facilitated. However, the knowledge of epidemiology and population diversity of MDR K. pneumoniae in KSA is scarce. We conducted a comprehensive genomic survey on 352 MDR K. pneumoniae isolates systematically collected from bloodstream and urinary tract infections in 34 hospitals across 15 major cities in KSA during 2022 and 2023. Whole-genome sequencing on the isolates was performed, followed by genomic epidemiology and phylodynamic analysis. Our study revealed a dynamic population characterized by the rapid expansion of several dominant clones, including, ST2096, ST147, and ST231, which were estimated to have emerged within the past decade. These clones exhibited widespread dissemination across hospitals and were genetically linked to global strains, particularly from the Middle East and South Asia. All major clones harboured plasmid-borne ESBLs and carbapenemase genes, with plasmidome analysis identifying multiple IncH, IncA/C and IncL plasmids underlying the MDR-hypervirulent phenotype. These plasmids were shared between major clones and became acquired on the same time scales as the expansion of the dominant clones. Our results report ST2096 as an emerging MDR-hypervirulent clone, emphasizing the need for monitoring of the circulating clones and their plasmid content in the KSA and broader West Asia.
Keywords: Klebsiella pneumoniae; antimicrobial resistance; hypervirulence and multidrug resistance (MDR); precision epidemiology; whole genome sequencing.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Holt KE, Wertheim H, Zadoks RN, et al. . Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. Jul 7 2015;112(27):E3574–E3581. doi:10.1073/pnas.1501049112 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
