Identification of potential hub genes associated with recurrent miscarriage through combined transcriptomic and proteomic analysis
- PMID: 39508749
- PMCID: PMC12042675
- DOI: 10.17305/bb.2024.11158
Identification of potential hub genes associated with recurrent miscarriage through combined transcriptomic and proteomic analysis
Abstract
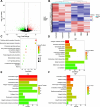
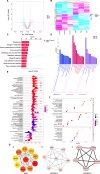
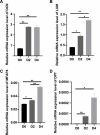
Recurrent miscarriage (RM) is currently difficult to prevent and treat due to a lack of comprehensive understanding of its molecular mechanisms. The aim of this study was to identify genes potentially involved in the pathogenesis of RM and to observe their expression in the decidual tissues of RM patients. A total of 1823 differentially expressed genes (DEGs) and 148 differentially expressed proteins (DEPs) in decidual tissues between RM and control groups were identified. Subsequently, DCN, DPT, LUM, MFAP4, and ISG15 were identified from the DEGs/DEPs as RM-related hub genes through systematic bioinformatics analysis. Bioinformatics analysis of the single-cell dataset GSE214607 revealed that the expression of these five hub genes in the decidual stromal cells (DSCs) of RM patients appeared to be upregulated, while the RT-qPCR assay showed that their decidual expression levels were significantly increased in RM patients. Uterine Isg15 expression was significantly increased, whereas the uterine expression of Dcn, Dpt, Lum, and Mfap4 was decreased in LPS-induced early pregnancy loss (EPL) mice. MiR-16-5p, -21-3p, -27a-3p, and -941 were identified as potentially involved in the regulation of these five hub genes, and their decidual expression levels were significantly decreased in RM patients. The abnormally increased ISG15 expression in the decidual tissues of RM patients and uterine tissues of LPS-induced mice was validated by WB analysis. ISG15 expression was significantly reduced during the in vitro decidualization of human endometrial stromal cells (hESCs). Collectively, DCN, DPT, LUM, MFAP4, and ISG15 were identified as RM-related hub genes, and their expression in the decidual tissues of RM patients was significantly increased. The decidualization of hESCs was accompanied by reduced ISG15 expression, suggesting that increased decidual ISG15 expression might lead to EPL by disrupting the decidualization process.
Conflict of interest statement
Conflicts of interest: Authors declare no conflicts of interest.
Figures





Similar articles
-
FXYD1 was identified as a hub gene in recurrent miscarriage and involved in decidualization via regulating Na/K-ATPase activity.J Assist Reprod Genet. 2025 Feb;42(2):665-678. doi: 10.1007/s10815-024-03363-8. Epub 2024 Dec 27. J Assist Reprod Genet. 2025. PMID: 39730944
-
Deep-sequencing identification of differentially expressed miRNAs in decidua and villus of recurrent miscarriage patients.Arch Gynecol Obstet. 2016 May;293(5):1125-35. doi: 10.1007/s00404-016-4038-5. Epub 2016 Feb 15. Arch Gynecol Obstet. 2016. PMID: 26879955 Free PMC article.
-
miR-3074-5p/CLN8 pathway regulates decidualization in recurrent miscarriage.Reproduction. 2021 May 27;162(1):33-45. doi: 10.1530/REP-21-0032. Reproduction. 2021. PMID: 34044364
-
The role of decidual cells in uterine hemostasis, menstruation, inflammation, adverse pregnancy outcomes and abnormal uterine bleeding.Hum Reprod Update. 2016 Jun;22(4):497-515. doi: 10.1093/humupd/dmw004. Epub 2016 Feb 23. Hum Reprod Update. 2016. PMID: 26912000 Free PMC article. Review.
-
Multiomics Studies Investigating Recurrent Pregnancy Loss: An Effective Tool for Mechanism Exploration.Front Immunol. 2022 Apr 27;13:826198. doi: 10.3389/fimmu.2022.826198. eCollection 2022. Front Immunol. 2022. PMID: 35572542 Free PMC article. Review.
References
-
- Quenby S, Gallos ID, Dhillon-Smith RK, Podesek M, Stephenson MD, Fisher J, et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet. 2021;397(10285):1658–67. https://doi.org/10.1016/S0140-6736(21)00682-6. - PubMed
-
- Pan HT, Ding HG, Fang M, Yu B, Cheng Y, Tan YJ, et al. Proteomics and bioinformatics analysis of altered protein expression in the placental villous tissue from early recurrent miscarriage patients. Placenta. 2018;61:1–10. https://doi.org/10.1016/j.placenta.2017.11.001. - PubMed
-
- Davalieva K, Terzikj M, Bozhinovski G, Kiprijanovska S, Kubelka-Sabit K, Plaseska-Karanfilska D. Comparative proteomics analysis of decidua reveals altered RNA processing and impaired ribosome function in recurrent pregnancy loss. Placenta. 2024;154:28–37. https://doi.org/10.1016/j.placenta.2024.06.005. - PubMed
-
- Sun F, Cui L, Qian J, Li M, Chen L, Chen C, et al. Decidual stromal cell Ferroptosis associated with abnormal iron metabolism is implicated in the pathogenesis of recurrent pregnancy loss. Int J Mol Sci. 2023;24(9):7836. https://doi.org/10.3390/ijms24097836. - PMC - PubMed
-
- Zhao X, Jiang Y, Luo S, Zhao Y, Zhao H. Intercellular communication involving macrophages at the maternal-fetal interface may be a pivotal mechanism of URSA: a novel discovery from transcriptomic data. Front Endocrinol (Lausanne) 2023;14:973930. https://doi.org/10.3389/fendo.2023.973930. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

