HIRA protects telomeres against R-loop-induced instability in ALT cancer cells
- PMID: 39509271
- PMCID: PMC11698518
- DOI: 10.1016/j.celrep.2024.114964
HIRA protects telomeres against R-loop-induced instability in ALT cancer cells
Erratum in
-
HIRA protects telomeres against R-loop-induced instability in ALT cancer cells.Cell Rep. 2025 Mar 25;44(3):115497. doi: 10.1016/j.celrep.2025.115497. Epub 2025 Mar 18. Cell Rep. 2025. PMID: 40106431 No abstract available.
Abstract
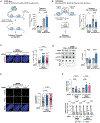
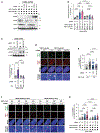
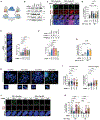
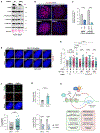
Inactivating mutations in chromatin modifiers, like the α-thalassemia/mental retardation, X-linked (ATRX)-death domain-associated protein (DAXX) chromatin remodeling/histone H3.3 deposition complex, drive the cancer-specific alternative lengthening of telomeres (ALT) pathway. Prior studies revealed that HIRA, another histone H3.3 chaperone, compensates for ATRX-DAXX loss at telomeres to sustain ALT cancer cell survival. How HIRA rescues telomeres from the consequences of ATRX-DAXX deficiency remains unclear. Here, using an assay for transposase-accessible chromatin using sequencing (ATAC-seq) and cleavage under targets and release using nuclease (CUT&RUN), we establish that HIRA-mediated deposition of new H3.3 maintains telomeric chromatin accessibility to prevent the detrimental accumulation of nucleosome-free single-stranded DNA (ssDNA) in ATRX-DAXX-deficient ALT cells. We show that the HIRA-UBN1/UBN2 complex deposits new H3.3 to prevent TERRA R-loop buildup and transcription-replication conflicts (TRCs) at telomeres. Furthermore, HIRA-mediated H3.3 incorporation into telomeric chromatin links productive ALT to the phosphorylation of serine 31, an H3.3-specific amino acid, by Chk1. Therefore, we identify a critical role for HIRA-mediated H3.3 deposition that ensures the survival of ATRX-DAXX-deficient ALT cancer cells.
Keywords: ALT; CP: Cancer; CP: Molecular biology; HIRA; R-loop; cancer; histone; telomere.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

