Odorant Binding Proteins Facilitate the Gas-Phase Uptake of Odorants Through the Nasal Mucus
- PMID: 39509459
- PMCID: PMC11724230
- DOI: 10.1002/chem.202403058
Odorant Binding Proteins Facilitate the Gas-Phase Uptake of Odorants Through the Nasal Mucus
Abstract
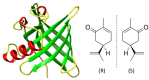
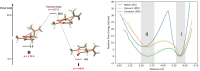
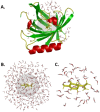
Mammalian odorant binding proteins (OBPs) have long been suggested to transport hydrophobic odorant molecules through the aqueous environment of the nasal mucus. While the function of OBPs as odorant transporters is supported by their hydrophobic beta-barrel structure, no rationale has been provided on why and how these proteins facilitate the uptake of odorants from the gas phase. Here, a multi-scale computational approach validated through available high-resolution spectroscopy experiments reveals that the conformational space explored by carvone inside the binding cavity of porcine OBP (pOBP) is much closer to the gas than the aqueous phase, and that pOBP effectively manages to transport odorants by lowering the free energy barrier of odorant uptake. Understanding such perireceptor events is crucial to fully unravel the molecular processes underlying the olfactory sense and move towards the development of protein-based biomimetic sensor units that can serve as artificial noses.
Keywords: Biosensors; Carvone; Enhanced Sampling; High-Resolution Spectroscopy; Odorant Binding.
© 2024 The Author(s). Chemistry - A European Journal published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

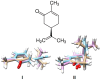
References
-
- Pelosi P., Knoll W., Biol. Rev. 2022, 97, 20. - PubMed
-
- Zaremska V., Tan J., Lim S., Knoll W., Pelosi P., Chem. Eur. J. 2020, 26, 8720. - PubMed
-
- Heydel J. M., Coelho A., Thiebaud N., Legendre A., Bon A. M. L., Faure P., Neiers F., Artur Y., Golebiowski J., Briand L., Anat. Rec. 2013, 296, 1333. - PubMed
-
- Spinelli S., Ramoni R., Grolli S., Bonicel J., Cambillau C., Tegoni M., Biochemistry 1998, 37, 7913. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

