Beyond IC50-A computational dynamic model of drug resistance in enzyme inhibition treatment
- PMID: 39509464
- PMCID: PMC11575782
- DOI: 10.1371/journal.pcbi.1012570
Beyond IC50-A computational dynamic model of drug resistance in enzyme inhibition treatment
Abstract
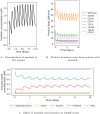
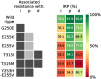
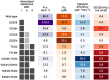
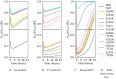
Resistance to therapy is a major clinical obstacle to treatment of cancer and communicable diseases. Drug selection in treatment of patients where the disease is showing resistance to therapy is often guided by IC50 or fold-IC50 values. In this work, through a model of the treatment of chronic myeloid leukaemia (CML), we contest using fold-IC50 values as a guide for treatment selection. CML is a blood cancer that is treated with Abl1 inhibitors, and is often seen as a model for targeted therapy and drug resistance. Resistance to the first-line treatment occurs in approximately one in four patients. The most common cause of resistance is mutations in the Abl1 enzyme. Different mutant Abl1 enzymes show resistance to different Abl1 inhibitors and the mechanisms that lead to resistance for various mutation and inhibitor combinations are not fully known, making the selection of Abl1 inhibitors for treatment a difficult task. We developed a model based on information of catalysis, inhibition and pharmacokinetics, and applied it to study the effect of three Abl1 inhibitors on mutants of the Abl1 enzyme. From this model, we show that the relative decrease of product formation rate (defined in this work as "inhibitory reduction prowess") is a better indicator of resistance than an examination of the size of the product formation rate or fold-IC50 values for the mutant. We also examine current ideas and practices that guide treatment choice and suggest a new parameter for selecting treatments that could increase the efficacy and thus have a positive impact on patient outcomes.
Copyright: © 2024 Roadnight Sheehan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

