Multi-platform biomarkers of response to an immune checkpoint inhibitor in the neoadjuvant I-SPY 2 trial for early-stage breast cancer
- PMID: 39510069
- PMCID: PMC11604542
- DOI: 10.1016/j.xcrm.2024.101799
Multi-platform biomarkers of response to an immune checkpoint inhibitor in the neoadjuvant I-SPY 2 trial for early-stage breast cancer
Abstract
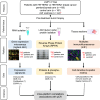
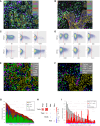
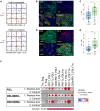
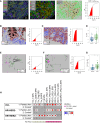
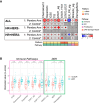
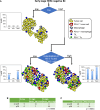
Only a subset of patients with breast cancer responds to immune checkpoint blockade (ICB). To better understand the underlying mechanisms, we analyze pretreatment biopsies from patients in the I-SPY 2 trial who receive neoadjuvant ICB using multiple platforms to profile the tumor microenvironment. A variety of immune cell populations and markers of immune/cytokine signaling associate with pathologic complete response (pCR). Interestingly, these differ by breast cancer receptor subtype. Measures of the spatial distributions of immune cells within the tumor microenvironment, in particular colocalization or close spatial proximity of PD-1+ T cells with PD-L1+ cells (immune and tumor cells), are significantly associated with response in the overall cohort as well as the in the triple negative (TN) and HR+HER2- subtypes. Our findings indicate that biomarkers associated with immune cell signaling, immune cell densities, and spatial metrics are predictive of neoadjuvant ICB efficacy in breast cancer.
Keywords: breast cancer; immune checkpoint blockade; multiplex immunofluorescence; predictive markers; spatial metrics.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.W. reports honoraria from DAVA Oncology; consults for Baylor College of Medicine; has ownership in Theralink; and is co-inventor of the RPPA technology, and phospho-HER2 and -EGFR response predictors with filed patents. C.H. is an employee of Akoya Biosciences. R.N. reports grants from Quantum Leap, AstraZeneca, Celgene, Corcept Therapeutics, Genentech/Roche, Immunomedics, Merck, OBI Pharma, Odonate Therapeutics, Pfizer, and Seattle Genetics outside the submitted work; and personal fees from Aduro, AstraZeneca, Athenex, Celgene, Daiichi Sankyo, G1 Therapeutics, Genentech, MacroGenics, Merck, Novartis, Pfizer, Puma, and Syndax. M.C.L. reports support from Eisai, Genentech, GRAIL, Menarini Silicon Biosystems, Merck, Novartis, Seattle Genetics, and Tesaro. D.Y. reports unrelated support from Boehringer Ingelheim. A.M.D. reports honoraria or consulting for Pfizer and Context Therapeutics and reports support from Novartis, Pfizer, Genentech, Calithera, and Menarini. L.P. reports consulting fees and honoraria for advisory board participation from Pfizer, AstraZeneca, Merck, Novartis, Bristol-Myers Squibb, GlaxoSmithKline, Genentech/Roche, Personalis, Daiichi, Natera, and Exact Sciences and institutional research funding from Seagen, GlaxoSmithKline, AstraZeneca, Merck, Pfizer, and Bristol-Myers Squibb. D.A.B. is co-owner of Berry Consultants LLC, a company that designs adaptive clinical trials (including I-SPY2). E.F.P. reports leadership, stock/ownership, consulting/advisory, and travel funds from Perthera and Ceres Nanosciences; stock and consulting/advisory for Theralink Technologies, Inc; support from Ceres Nanosciences, GlaxoSmithKline, AbbVie, Symphogen, Genentech, SpringWorks Therapeutics, and Deciphera Therapeutics; and patents/royalties from NIH and filed patents for p-HER2 and p-EGFR response predictors. L.v.V. is a co-inventor of the MammaPrint signature and a part-time employee and stockholder of Agendia NV. L.E. is an unpaid member of the board of directors of Quantum Leap Healthcare Collaborative (QLHC), has received grant support from QLHC for the I-SPY2 trial, and is on the Blue Cross/Blue Shield Medical Advisory Panel.
Figures


References
-
- Burtness B., Harrington K.J., Greil R., Soulières D., Tahara M., de Castro G., Psyrri A., Basté N., Neupane P., Bratland Å., et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. 2019;394:1915–1928. doi: 10.1016/S0140-6736(19)32591-7. - DOI - PubMed
-
- Motzer R.J., Tannir N.M., McDermott D.F., Arén Frontera O., Melichar B., Choueiri T.K., Plimack E.R., Barthélémy P., Porta C., George S., et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2018;378:1277–1290. doi: 10.1056/NEJMoa1712126. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

