Legius syndrome mutations in the Ras-regulator SPRED1 abolish its membrane localization and potentially cause neurodegeneration
- PMID: 39510187
- PMCID: PMC11648228
- DOI: 10.1016/j.jbc.2024.107969
Legius syndrome mutations in the Ras-regulator SPRED1 abolish its membrane localization and potentially cause neurodegeneration
Abstract
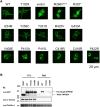
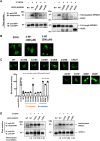
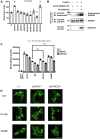
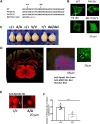
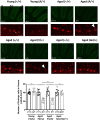
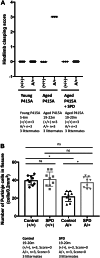
The SPRED family proteins act as negative regulators of the Ras-ERK pathway: the N-terminal EVH1 domain interacts with the Ras-GAP domain (GRD) of the NF1 protein, while the C-terminal Sprouty-related (SPR) domain promotes membrane localization of SPRED, thereby recruiting NF-1 to Ras. Loss-of-function mutations in the hSPRED1 cause Legius syndrome in an autosomal dominant manner. In this study, we investigated the effects of missense mutations in the SPR domain identified in patients with Legius syndrome. Among the 18 mutations we examined, six (C368S, M369L, V408E, P415A, P415L, and P422R) have defects in the palmitoylation of the SPRED1 protein, losing plasma membrane localization and forming cytoplasmic granular aggregates. To evaluate the in vivo effects of SPR mutations, knock-in (KI) mice with P415A and P415V substitutions or M417Afs∗4, a C-terminal 28 amino acid deletion, were generated. All these KI mice exhibited cranial malformations, a characteristic feature of Legius syndrome. However, both P415A and P415V mutants formed granular aggregates, whereas M417Afs∗4 showed a diffuse cytoplasmic distribution, and Spred1P415A and Spred1P415V mice, but not Spred1M417Afs∗4 mice, developed cerebellar ataxia and Purkinje cell loss with age. These data suggest that in addition to loss of palmitoylation, the C-terminal region is required for the granular aggregate formation and Purkinje cell loss. The autophagy inducer spermidine rescued the ataxia phenotypes and Purkinje cell loss in Spred1P415A mice. These results suggest that some, but not all, SPR mutations that lose lipid modification induce abnormal cytoplasmic aggregation, which could be a target for autophagic clearance, and potentially cause neurodegenerative diseases.
Keywords: Legius syndrome; SPRED1; autophagy; membrane localization; neurodegeneration; palmitoylation; spermidine.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Brems H., Legius E. Legius syndrome, an update. Molecular pathology of mutations in SPRED1. Keio J Med. 2013;62:107–112. - PubMed
-
- Messiaen L., Yao S., Brems H., Callens T., Sathienkijkanchai A., Denayer E., et al. Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA. 2009;302:2111–2118. - PubMed
-
- Legius E., Stevenson D. In: GeneReviews(®) Adam M.P., Feldman J., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., et al., editors. University of Washington, Seattle; Seattle, WA: 1993. Legius Syndrome. - PubMed
-
- Pasmant E., Gilbert-Dussardier B., Petit A., de Laval B., Luscan A., Gruber A., et al. SPRED1, a RAS MAPK pathway inhibitor that causes Legius syndrome, is a tumour suppressor downregulated in paediatric acute myeloblastic leukaemia. Oncogene. 2015;34:631–638. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

