Mass spectrometry-based proteomic exploration of diverse murine macrophage cellular models
- PMID: 39510801
- PMCID: PMC11544424
- DOI: 10.26508/lsa.202402760
Mass spectrometry-based proteomic exploration of diverse murine macrophage cellular models
Abstract
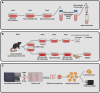
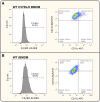
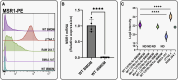
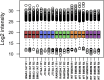
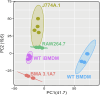
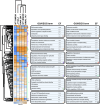
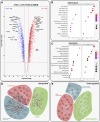
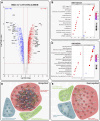
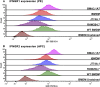
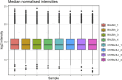
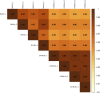
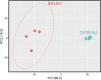
Immortalised cell lines that mimic their primary cell counterparts are fundamental to research, particularly when large cell numbers are required. Here, we report that immortalisation of bone marrow-derived macrophages (iBMDMs) using the J2 virus resulted in the loss of a protein of interest, MSR1, in WT cells by an unknown mechanism. This led us to perform an in-depth mass spectrometry-based proteomic characterisation of common murine macrophage cell lines (J774A.1, RAW264.7, and BMA3.1A7), in comparison with the iBMDMs, as well as primary BMDMs from both C57BL/6 and BALB/c mice. This analysis revealed striking differences in protein profiles associated with macrophage polarisation, phagocytosis, pathogen recognition, and interferon signalling. Among the cell lines, J774A.1 cells were the most similar to the gold standard primary BMDM model, whereas BMA3.1A7 cells were the least similar because of the reduction in abundance of several key proteins related closely to macrophage function. This comprehensive proteomic dataset offers valuable insights into the use and suitability of macrophage cell lines for cell signalling and inflammation research.
© 2024 Gudgeon et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Assouvie A, Daley-Bauer LP, Rousselet G (2018) Growing murine bone marrow-derived macrophages. In Macrophages: Methods and Protocols, Rousselet G (ed.), pp 29–33. New York, NY: Springer. - PubMed
-
- Banete A, Achita P, Harding K, Mulder R, Basta S (2015) Immortalized murine macrophage cell line as a model for macrophage polarization into classically activated M(IFNγ+LPS) or alternatively activated M(IL-4) macrophages. J Clin Cell Immunol 6: 318. 10.4172/2155-9899.1000318 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
