TET2 germline variants promote kidney disease by impairing DNA repair and activating cytosolic nucleotide sensors
- PMID: 39511169
- PMCID: PMC11543665
- DOI: 10.1038/s41467-024-53798-x
TET2 germline variants promote kidney disease by impairing DNA repair and activating cytosolic nucleotide sensors
Abstract
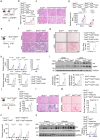
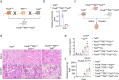
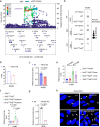
Genome-wide association studies (GWAS) have identified over 800 loci associated with kidney function, yet the specific genes, variants, and pathways involved remain elusive. By integrating kidney function GWAS with human kidney expression and methylation quantitative trait analyses, we identified Ten-Eleven Translocation (TET) DNA demethylase 2 (TET2) as a novel kidney disease risk gene. Utilizing single-cell chromatin accessibility and CRISPR-based genome editing, we highlight GWAS variants that influence TET2 expression in kidney proximal tubule cells. Experiments using kidney/tubule-specific Tet2 knockout mice indicated its protective role in cisplatin-induced acute kidney injury, as well as in chronic kidney disease and fibrosis induced by unilateral ureteral obstruction or adenine diet. Single-cell gene profiling of kidneys from Tet2 knockout mice and TET2-knockdown tubule cells revealed the altered expression of DNA damage repair and chromosome segregation genes, notably including INO80, another kidney function GWAS target gene itself. Remarkably, both TET2-null and INO80-null cells exhibited an increased accumulation of micronuclei after injury, leading to the activation of cytosolic nucleotide sensor cGAS-STING. Genetic deletion of cGAS or STING in kidney tubules, or pharmacological inhibition of STING, protected TET2-null mice from disease development. In conclusion, our findings highlight TET2 and INO80 as key genes in the pathogenesis of kidney diseases, indicating the importance of DNA damage repair mechanisms.
© 2024. The Author(s).
Conflict of interest statement
The Susztak lab is supported by Boehringer Ingelheim, Regeneron, Bayer, GSK, Gilead, Jnana, Maze, Novartis, ONO and Novo Nordisk for work that is unrelated to the current manuscript. The remaining authors declare no competing interests.
Figures




References
-
- Jager, K. J. et al. A single number for advocacy and communication-worldwide more than 850 million individuals have kidney diseases. Nephrol. Dial. Transpl.34, 1803–1805 (2019). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R01DK076077/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01DK076077/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01DK132630/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01DK105821/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01DK087635/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

