Altered genomic methylation promotes Staphylococcus aureus persistence in hospital environment
- PMID: 39511195
- PMCID: PMC11544029
- DOI: 10.1038/s41467-024-54033-3
Altered genomic methylation promotes Staphylococcus aureus persistence in hospital environment
Abstract
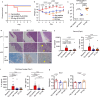
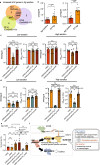
Staphylococcus aureus can cause outbreaks and becomes multi-drug resistant through gene mutations and acquiring resistance genes. However, why S. aureus easily adapts to hospital environments, promoting resistance and recurrent infections, remains unknown. Here we show that a specific S. aureus lineage evolved from a clone that expresses the accessory gene regulator (Agr) system to subclones that reversibly suppressed Agr and caused an outbreak in the hospital setting. S. aureus with flexible Agr regulation shows increased ability to acquire antibiotic-resistant plasmids, escape host immunity, and colonize mice. Bacteria with flexible Agr regulation shows altered cytosine genomic methylation, including the decreased 5mC methylation in transcriptional regulator genes (pcrA and rpsD), compared to strains with normal Agr expression patterns. In this work, we discover how altered genomic methylation promotes flexible Agr regulation which is associated with persistent pathogen colonization in the hospital environment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Naimi, T. S. et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA290, 2976–2984 (2003). - PubMed
-
- Liu, J., Gefen, O., Ronin, I., Bar-Meir, M. & Balaban, N. Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science367, 200–204 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 18gm6010016h0002/Japan Agency for Medical Research and Development (AMED)
- 23gm1610004h0003/Japan Agency for Medical Research and Development (AMED)
- 23gm1610004h0003/Japan Agency for Medical Research and Development (AMED)
- 23gm1610004h0003/Japan Agency for Medical Research and Development (AMED)
- ZIA-AI000904/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
LinkOut - more resources
Full Text Sources
Medical

