Mapping membrane biophysical nano-environments
- PMID: 39511199
- PMCID: PMC11544141
- DOI: 10.1038/s41467-024-53883-1
Mapping membrane biophysical nano-environments
Abstract
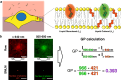
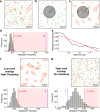
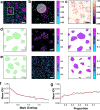
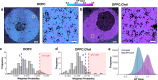
The mammalian plasma membrane is known to contain domains with varying lipid composition and biophysical properties. However, studying these membrane lipid domains presents challenges due to their predicted morphological similarity to the bulk membrane and their scale being below the classical resolution limit of optical microscopy. To address this, we combine the solvatochromic probe di-4-ANEPPDHQ, which reports on its biophysical environment through changes in its fluorescence emission, with spectrally resolved single-molecule localisation microscopy. The resulting data comprises nanometre-precision localisation coordinates and a generalised polarisation value related to the probe's environment - a marked point pattern. We introduce quantification algorithms based on topological data analysis (PLASMA) to detect and map nano-domains in this marked data, demonstrating their effectiveness in both artificial membranes and live cells. By leveraging environmentally sensitive fluorophores, multi-modal single molecule localisation microscopy, and advanced analysis methods, we achieve nanometre scale mapping of membrane properties and assess changes in response to external perturbation with methyl-β-cyclodextrin. This integrated methodology represents an integrated toolset for investigating marked point pattern data at nanometre spatial scales.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Klymchenko, A. S. Solvatochromic and fluorogenic dyes as environment-sensitive probes: design and biological applications. Acc. Chem. Res50, 366–375 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

