HuD impairs neuromuscular junctions and induces apoptosis in human iPSC and Drosophila ALS models
- PMID: 39511225
- PMCID: PMC11544248
- DOI: 10.1038/s41467-024-54004-8
HuD impairs neuromuscular junctions and induces apoptosis in human iPSC and Drosophila ALS models
Abstract
Defects at the neuromuscular junction (NMJ) are among the earliest hallmarks of amyotrophic lateral sclerosis (ALS). According to the "dying-back" hypothesis, NMJ disruption not only precedes but also triggers the subsequent degeneration of motoneurons in both sporadic (sALS) and familial (fALS) ALS. Using human induced pluripotent stem cells (iPSCs), we show that the RNA-binding protein HuD (ELAVL4) contributes to NMJ defects and apoptosis in FUS-ALS. HuD overexpression mimics the severe FUSP525L mutation, while its knockdown rescues the FUSP525L phenotypes. In Drosophila, neuronal overexpression of the HuD ortholog, elav, induces motor dysfunction, and its knockdown improves motor function in a FUS-ALS model. Finally, we report increased HuD levels upon oxidative stress in human motoneurons and in sALS patients with an oxidative stress signature. Based on these findings, we propose that HuD plays a role downstream of FUS mutations in fALS and in sALS related to oxidative stress.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


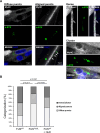

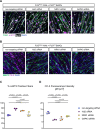

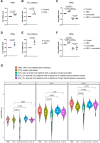
References
-
- Brown, R. H. & Al-Chalabi, A. Amyotrophic lateral sclerosis. N. Engl. J. Med.377, 162–172 (2017). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- CN00000041/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- 2022BYB33L/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- CN00000041/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- RP123188EC9F2349/Sapienza Università di Roma (Sapienza University of Rome)
- RM12117A5DE7A45B/Sapienza Università di Roma (Sapienza University of Rome)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

