PTGS is dispensable for the initiation of epigenetic silencing of an active transposon in Arabidopsis
- PMID: 39511423
- PMCID: PMC11624286
- DOI: 10.1038/s44319-024-00304-5
PTGS is dispensable for the initiation of epigenetic silencing of an active transposon in Arabidopsis
Abstract
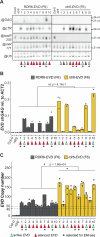
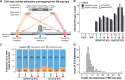
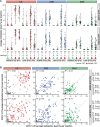
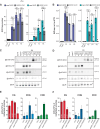
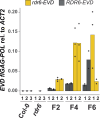
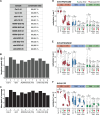
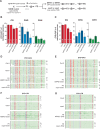
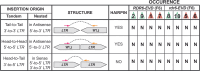
Transposable elements (TEs) are repressed in plants through transcriptional gene silencing (TGS), maintained epigenetic silencing marks such as DNA methylation. However, the mechanisms by which silencing is first installed remain poorly understood in plants. Small interfering (si)RNAs and post-transcriptional gene silencing (PTGS) are believed to mediate the initiation of TGS by guiding the first deposition of DNA methylation. To determine how this silencing installation works, we took advantage of ÉVADÉ (EVD), an endogenous retroelement in Arabidopsis, able to recapitulate true de novo silencing with a sequence of PTGS followed by a TGS. To test whether PTGS is required for TGS, we introduce active EVD into RNA-DEPENDENT-RNA-POLYMERASE-6 (RDR6) mutants, an essential PTGS component. EVD activity and silencing are monitored across several generations. In the absence of PTGS, silencing of EVD is still achieved through installation of RNA-directed DNA methylation (RdDM). Our study shows that PTGS is dispensable for de novo EVD silencing. Although we cannot rule out that PTGS might facilitate TGS, or control TE activity, initiation of epigenetic silencing can take place in its absence.
Keywords: Epigenetics; Plants; Silencing; Transposons; siRNAs.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures




References
-
- Al-Kaff NS, Covey SN, Kreike MM, Page AM, Pinder R, Dale PJ (1998) Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113–2115 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657 - PubMed
-
- Baduel P, Quadrana L, Colot V (2021) Plant transposable elements, methods and protocols. Methods Mol Biol 2250:157–169 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

