Collaboration between clinicians and vision-language models in radiology report generation
- PMID: 39511432
- PMCID: PMC11835717
- DOI: 10.1038/s41591-024-03302-1
Collaboration between clinicians and vision-language models in radiology report generation
Abstract
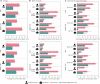
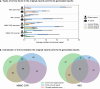
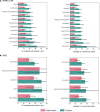
Automated radiology report generation has the potential to improve patient care and reduce the workload of radiologists. However, the path toward real-world adoption has been stymied by the challenge of evaluating the clinical quality of artificial intelligence (AI)-generated reports. We build a state-of-the-art report generation system for chest radiographs, called Flamingo-CXR, and perform an expert evaluation of AI-generated reports by engaging a panel of board-certified radiologists. We observe a wide distribution of preferences across the panel and across clinical settings, with 56.1% of Flamingo-CXR intensive care reports evaluated to be preferable or equivalent to clinician reports, by half or more of the panel, rising to 77.7% for in/outpatient X-rays overall and to 94% for the subset of cases with no pertinent abnormal findings. Errors were observed in human-written reports and Flamingo-CXR reports, with 24.8% of in/outpatient cases containing clinically significant errors in both report types, 22.8% in Flamingo-CXR reports only and 14.0% in human reports only. For reports that contain errors we develop an assistive setting, a demonstration of clinician-AI collaboration for radiology report composition, indicating new possibilities for potential clinical utility.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: This study was funded by Google LLC and/or a subsidiary thereof (‘Google’). R.T., D.G.T.B., A.Sellergren., S.G., S.D., A.See., J.W., C.L., T.T., S.A., M.S., R.M., R.L., S.Man., Z.A., S.Mahdavi., Y.M., J.B., S.M.A.E., Y.L., S.S., V.N., P.K., P.S.-H., A.K. and I.K. are employees of Google and may own stock as part of the standard compensation package. D.B. was a Google employee and is currently an employee of GlaxoSmithKlein AI division and may own stock as part of the standard compensation package. Similarly, K.S. was a Google employee and may own stock, but is currently an employee of OpenAI.
Figures








References
-
- Rimmer, A. Radiologist shortage leaves patient care at risk, warns Royal College. BMJ359, j4683 (2017). - PubMed
-
- Rajpurkar, P. & Lungren, M. P. The current and future state of AI interpretation of medical images. N. Engl. J. Med.388, 1981–1990 (2023). - PubMed
-
- Allen, B., Agarwal, S., Coombs, L., Wald, C. & Dreyer, K. 2020 ACR Data Science Institute artificial intelligence survey. J. Am. Coll. Radiol.18, 1153–1159 (2021). - PubMed
-
- Milam, M. E. & Koo, C. W. The current status and future of FDA-approved artificial intelligence tools in chest radiology in the United States. Clin. Radiol.78, 115–122 (2023). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

