Targeting LLT1 as a potential immunotherapy option for cancer patients non-responsive to existing checkpoint therapies in multiple solid tumors
- PMID: 39511540
- PMCID: PMC11545609
- DOI: 10.1186/s12885-024-13074-z
Targeting LLT1 as a potential immunotherapy option for cancer patients non-responsive to existing checkpoint therapies in multiple solid tumors
Abstract
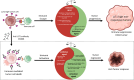
Background: High levels of LLT1 expression have been found in several cancers, where it interacts with CD161 on NK cells to facilitate tumor immune escape. Targeting LLT1 could potentially relieve this inhibitory signal and enhance anti-tumor responses mediated through NK cells. Using the 'The Cancer Genome Atlas' (TCGA) database, we investigated the role of LLT1 in the tumor microenvironment (TME) across various cancers. Identifying such biomarkers could create new therapeutic options for patients in addition to complementing existing immunotherapies.
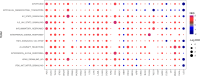
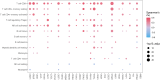
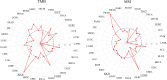
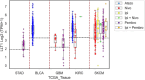
Methods: LLT1 expression was evaluated in 33 cancers using TCGA transcriptome data. Univariate Cox regression analysis was employed to assess the correlation of LLT1 expression with patient survival. The relationship between LLT1 expression with immune infiltrates, immune gene signatures, and cancer genomic biomarkers (TMB, MSI, and MMR) was also investigated. Immunofluorescence studies were conducted to validate LLT1 expression in tumors. Furthermore, using the CRI iAtlas data, we evaluated LLT1 distribution and its correlation with other immune checkpoint genes in patients non-responsive to existing immune checkpoint therapies across multiple solid cancers.
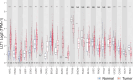
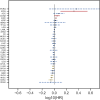
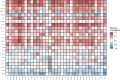
Results: High expression of LLT1 was observed in 12 cancers, including BRCA, CHOL, ESCA, GBM, HNSC, KIRC, KIRP, LIHC, LUAD, STAD, SARC, and PCPG. In certain cancers like COAD, KICH, and KIRC, high LLT1 expression was associated with poor prognosis. Further analysis revealed that upregulated LLT1 was associated with an abundance of NK and T cell infiltrates in the TME, as well as exhaustive immune biomarkers, and inversely associated with pro-inflammatory and tumor suppressor signatures. High LLT1 expression is also positively correlated with genomic biomarkers in certain cancers. Immunofluorescence studies confirmed moderate to high LLT1 expression in immune-resistant prostate cancer, glioma, ovarian cancer, and immune-sensitive liver cancer cell lines. An independent assessment of clinical cohorts from CRI iAtlas showed a correlation of upregulated LLT1 with multiple immunosuppressive genes in patients non-responsive to current ICIs.
Conclusions: The biomarker analysis revealed a clear association between elevated LLT1 expression and an immunosuppressive TME in patient cohorts from TCGA and clinical databases. Therefore, this study provides a foundation for utilizing LLT1 as a potential target to improve clinical responses in ICI non-responsive patients with upregulated LLT1.
Keywords: Immune Checkpoint Inhibitors; Immunotherapy; LLT1; Natural Killer Cells; Tumor Biomarkers; Tumor Microenvironment.
© 2024. The Author(s).
Conflict of interest statement
TM, SK, AT, AD, SB, YM, and SK are employees of Zumutor Biologics. SG is a former employee of Zumutor Biologics. MG, AT, AD, SB, and YM have stock options in Zumutor Biologics, the company that owns the anti-LLT1 antibody, ZM008. MSM and GR declared no competing interests.
Figures












References
-
- Boles KS, Barten R, Kumaresan PR, Trowsdale J, Mathew PA. Cloning of a new lectin-like receptor expressed on human NK cells. Immunogenetics. 1999;50(1–2):1–7. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

