Widespread 3'UTR capped RNAs derive from G-rich regions in proximity to AGO2 binding sites
- PMID: 39511645
- PMCID: PMC11546257
- DOI: 10.1186/s12915-024-02032-7
Widespread 3'UTR capped RNAs derive from G-rich regions in proximity to AGO2 binding sites
Abstract
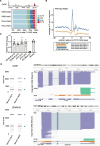
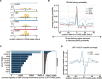
The 3' untranslated region (3'UTR) plays a crucial role in determining mRNA stability, localisation, translation and degradation. Cap analysis of gene expression (CAGE), a method for the detection of capped 5' ends of mRNAs, additionally reveals a large number of apparently 5' capped RNAs derived from locations within the body of the transcript, including 3'UTRs. Here, we provide direct evidence that these 3'UTR-derived RNAs are indeed capped and widespread in mammalian cells. By using a combination of AGO2 enhanced individual nucleotide resolution UV crosslinking and immunoprecipitation (eiCLIP) and CAGE following siRNA treatment, we find that these 3'UTR-derived RNAs likely originate from AGO2-binding sites, and most often occur at locations with G-rich motifs bound by the RNA-binding protein UPF1. High-resolution imaging and long-read sequencing analysis validate several 3'UTR-derived RNAs, showcase their variable abundance and show that they may not co-localise with the parental mRNAs. Taken together, we provide new insights into the origin and prevalence of 3'UTR-derived RNAs, show the utility of CAGE-seq for their genome-wide detection and provide a rich dataset for exploring new biology of a poorly understood new class of RNAs.
Keywords: 3′UTR; 3′UTR-derived RNA; AGO2; CAGE; Capping; G-rich; Subcellular localisation; UPF1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Naeli P, Winter T, Hackett AP, Alboushi L, Jafarnejad SM. The intricate balance between microRNA-induced mRNA decay and translational repression. FEBS J. 2023;290(10):2508–24. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

