The impact of cannabinoid receptor 1 absence on mouse liver mitochondria homeostasis: insight into mitochondrial unfolded protein response
- PMID: 39512900
- PMCID: PMC11541708
- DOI: 10.3389/fcell.2024.1464773
The impact of cannabinoid receptor 1 absence on mouse liver mitochondria homeostasis: insight into mitochondrial unfolded protein response
Abstract
Introduction: The contribution of Cannabinoid type 1 receptor (CB1) in mitochondrial energy transduction mechanisms and mitochondrial activities awaits deeper investigations. Our study aims to assess the impact of CB1 absence on the mitochondrial compartment in the liver, focusing on both functional aspects and remodeling processes.
Methods: We used CB1-/- and CB1+/+ male mice. Cytochrome C Oxidase activity was determined polarographically. The expression and the activities of separated mitochondrial complexes and supercomplexes were performed by using Blue-Native Page, Western blotting and histochemical staining for in-gel activity. Key players of Mitochondrial Quality Control processes were measured using RT-qPCR and Western blotting. Liver fine sub-cellular ultrastructural features were analyzed by TEM analysis.
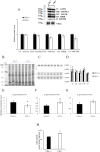
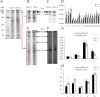
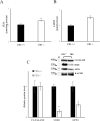
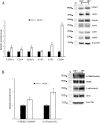
Results and discussion: In the absence of CB1, several changes in the liver occur, including increased oxidative capacity, reduced complex I activity, enhanced complex IV activity, general upregulation of respiratory supercomplexes, as well as higher levels of oxidative stress. The mitochondria and cellular metabolism may be affected by these changes, increasing the risk of ROS-related damage. CB1-/- mice show upregulation of mitochondrial fusion, fission and biogenesis processes which suggests a dynamic response to the absence of CB1. Furthermore, oxidative stress disturbs mitochondrial proteostasis, initiating the mitochondrial unfolded protein response (UPRmt). We noted heightened levels of pivotal enzymes responsible for maintaining mitochondrial integrity, along with heightened expression of molecular chaperones and transcription factors associated with cellular stress reactions. Additionally, our discoveries demonstrate a synchronized reaction to cellular stress, involving both UPRmt and UPRER pathways.
Keywords: cannabinoid receptor 1; homeostasis; mitochondrial quality control; mitochondrial unfolded protein response; oxidative stress; respiratory chain supercomplexes.
Copyright © 2024 Senese, Petito, Silvestri, Ventriglia, Mosca, Potenza, Russo, Falvo, Manfrevola, Cobellis, Chioccarelli, Porreca, Mele, Chianese, de Lange, Ricci, Cioffi and Lanni.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Effect of CB1 Receptor Deficiency on Mitochondrial Quality Control Pathways in Gastrocnemius Muscle.Biology (Basel). 2024 Feb 11;13(2):116. doi: 10.3390/biology13020116. Biology (Basel). 2024. PMID: 38392333 Free PMC article.
-
FUNDC1 activates the mitochondrial unfolded protein response to preserve mitochondrial quality control in cardiac ischemia/reperfusion injury.Cell Signal. 2022 Apr;92:110249. doi: 10.1016/j.cellsig.2022.110249. Epub 2022 Jan 17. Cell Signal. 2022. PMID: 35051611
-
Dysregulation of the Mitochondrial Unfolded Protein Response Induces Non-Apoptotic Dopaminergic Neurodegeneration in C. elegans Models of Parkinson's Disease.J Neurosci. 2017 Nov 15;37(46):11085-11100. doi: 10.1523/JNEUROSCI.1294-17.2017. Epub 2017 Oct 13. J Neurosci. 2017. PMID: 29030433 Free PMC article.
-
Mitochondria Retrograde Signaling and the UPR mt: Where Are We in Mammals?Int J Mol Sci. 2015 Aug 6;16(8):18224-51. doi: 10.3390/ijms160818224. Int J Mol Sci. 2015. PMID: 26258774 Free PMC article. Review.
-
Insight into the mitochondrial unfolded protein response and cancer: opportunities and challenges.Cell Biosci. 2022 Feb 18;12(1):18. doi: 10.1186/s13578-022-00747-0. Cell Biosci. 2022. PMID: 35180892 Free PMC article. Review.
Cited by
-
Dysfunctional Mitochondria Characterize Amyotrophic Lateral Sclerosis Patients' Cells Carrying the p.G376D TARDBP Pathogenetic Substitution.Antioxidants (Basel). 2025 Mar 28;14(4):401. doi: 10.3390/antiox14040401. Antioxidants (Basel). 2025. PMID: 40298692 Free PMC article.
References
-
- Aquila S., Guido C., Santoro A., Perrotta I., Laezza C., Bifulco M., et al. (2010). Human sperm anatomy: ultrastructural localization of the cannabinoid1 receptor and a potential role of anandamide in sperm survival and acrosome reaction. Anat. Rec. Hob. 293, 298–309. 10.1002/ar.21042 - DOI - PubMed
LinkOut - more resources
Full Text Sources

