Efficient PHB2 (prohibitin 2) exposure during mitophagy depends on VDAC1 (voltage dependent anion channel 1)
- PMID: 39513197
- PMCID: PMC12506749
- DOI: 10.1080/15548627.2024.2426116
Efficient PHB2 (prohibitin 2) exposure during mitophagy depends on VDAC1 (voltage dependent anion channel 1)
Abstract
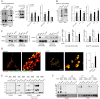
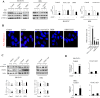
Exposure of inner mitochondrial membrane resident protein PHB2 (prohibitin 2) during autophagic removal of depolarized mitochondria (mitophagy) depends on the ubiquitin-proteasome system. This uncovering facilitates the PHB2 interaction with phagophore membrane-associated protein MAP1LC3/LC3. It is unclear whether PHB2 is exposed randomly at mitochondrial rupture sites. Prior knowledge and initial screening indicated that VDAC1 (voltage dependent anion channel 1) might play a role in this phenomenon. Through in vitro biochemical assays and imaging, we have found that VDAC1-PHB2 interaction increases during mitochondrial depolarization. Subsequently, this interaction enhances the efficiency of PHB2 exposure and mitophagy. To investigate the relevance in vivo, we utilized porin (equivalent to VDAC1) knockout Drosophila line. Our findings demonstrate that during mitochondrial stress, porin is essential for Phb2 exposure, Phb2-Atg8 interaction and mitophagy. This study highlights that VDAC1 predominantly synchronizes efficient PHB2 exposure through mitochondrial rupture sites during mitophagy. These findings may provide insights to understand progressive neurodegeneration.
Keywords: Neurodegeneration; PHB2-LC3 interaction; PINK1-PRKN; parkinson disease; porin; ubiquitin-proteasome system.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
