Myocardial Posttranscriptional Landscape in Peripartum Cardiomyopathy
- PMID: 39513265
- PMCID: PMC11643137
- DOI: 10.1161/CIRCHEARTFAILURE.124.011725
Myocardial Posttranscriptional Landscape in Peripartum Cardiomyopathy
Abstract
Background: Pregnancy imposes significant cardiovascular adaptations, including progressive increases in plasma volume and cardiac output. For most women, this physiological adaptation resolves at the end of pregnancy, but some women develop pathological dilatation and ultimately heart failure late in pregnancy or in the postpartum period, manifesting as peripartum cardiomyopathy (PPCM). Despite the mortality risk of this form of heart failure, the molecular mechanisms underlying PPCM have not been extensively examined in human hearts.
Methods: Protein and metabolite profiles from left ventricular tissue of end-stage PPCM patients (N=6-7) were compared with dilated cardiomyopathy (DCM; N=5-6) and nonfailing donors (N=7-18) using unbiased quantitative mass spectrometry. All samples were derived from the Sydney Heart Bank. Data are available via ProteomeXchange with identifier PXD055986. Differential protein expression and metabolite abundance and Kyoto Encyclopedia of Genes and Genomes pathway analyses were performed.
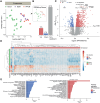
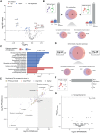
Results: Proteomic analysis identified 2 proteins, SBSPON (somatomedin B and thrombospondin type 1 domain-containing protein precursor) and TNS3 (tensin 3), that were uniquely downregulated in PPCM. SBSPON, an extracellular matrix protein, and TNS3, involved in actin remodeling and cell signaling, may contribute to impaired tissue remodeling and fibrosis in PPCM. Metabolomic analysis revealed elevated levels of homogentisate and deoxycholate and reduced levels of lactate and alanine in PPCM, indicating disrupted metabolic pathways and glucose utilization. Both PPCM and DCM shared pathways related to inflammation, immune responses, and signal transduction. However, thyroid hormone signaling was notably reduced in PPCM, affecting contractility and calcium handling through altered expression of PLN (phospholamban) and Sarcoendoplasmic Reticulum Calcium ATPase (SERCA). Enhanced endoplasmic reticulum stress and altered endocytosis pathways in PPCM suggested additional mechanisms of energy metabolism disruption.
Conclusions: The present study reveals unique posttranslational molecular features of the PPCM myocardium, which mediates cellular and metabolic remodeling, and holds promise as potential targets for therapeutic intervention.
Keywords: cardiomyopathy; heart failure; metabolomics; myocardium; peripartum period; pregnancy; proteomics.
Conflict of interest statement
None.
Figures




References
-
- Ferreira AF, Azevedo MJ, Proenca T, Saraiva FA, Machado AP, Sousa C, Sampaio Maia B, Leite-Moreira A, Ramalho C, Fa I. Cardiac remodelling and reverse remodelling in pregnant women: what can be expected? Eur Heart J. 2021;42:ehab724.2904. doi: 10.1093/eurheartj/ehab724.2904
-
- Sliwa K, Bauersachs J, Arany Z, Spracklen TF, Hilfiker-Kleiner D. Peripartum cardiomyopathy: from genetics to management. Eur Heart J. 2021;42:3094–3102. doi: 10.1093/eurheartj/ehab458 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

