Inhibition of Bruton's tyrosine kinase with PD-1 blockade modulates T cell activation in solid tumors
- PMID: 39513363
- PMCID: PMC11601564
- DOI: 10.1172/jci.insight.169927
Inhibition of Bruton's tyrosine kinase with PD-1 blockade modulates T cell activation in solid tumors
Abstract
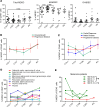
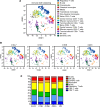
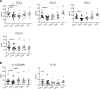
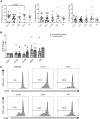
BACKGROUNDInhibition of Bruton's tyrosine kinase with ibrutinib blocks the function of myeloid-derived suppressor cells (MDSC). The combination of ibrutinib and nivolumab was tested in patients with metastatic solid tumors.METHODSSixteen patients received ibrutinib 420 mg p.o. daily with nivolumab 240 mg i.v. on days 1 and 15 of a 28-day cycle. The effect of ibrutinib and nivolumab on MDSC, the immune profile, and cytokine levels were measured. Single-cell RNA-Seq and T cell receptor sequencing of immune cells was performed.RESULTSCommon adverse events were fatigue and anorexia. Four patients had partial responses and 4 had stable disease at 3 months (average 6.5 months, range 3.5-14.6). Median overall survival (OS) was 10.8 months. Seven days of Bruton's tyrosine kinase (BTK) inhibition significantly increased the proportion of monocytic-MDSC (M-MDSC) and significantly decreased chemokines associated with MDSC recruitment and accumulation (CCL2, CCL3, CCL4, CCL13). Single-cell RNA-Seq revealed ibrutinib-induced downregulation of genes associated with MDSC-suppressive function (TIMP1, CXCL8, VEGFA, HIF1A), reduced MDSC interactions with exhausted CD8+ T cells, and decreased TCR repertoire diversity. The addition of nivolumab significantly increased circulating NK and CD8+ T cells and increased CD8+ T cell proliferation. Exploratory analyses suggest that MDSC and T cell gene expression and TCR repertoire diversity were differentially affected by BTK inhibition according to patient response.CONCLUSIONIbrutinib and nivolumab were well tolerated and affected MDSC and T cell function in patients with solid metastatic tumors.TRIAL REGISTRATIONClinicalTrials.gov NCT03525925.FUNDINGNIH; National Cancer Institute Cancer; National Center for Advancing Translational Sciences; Pelotonia.
Keywords: Cancer immunotherapy; Cellular immune response; Clinical trials; Immunology; T cells.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

