Diversity of the immune microenvironment and response to checkpoint inhibitor immunotherapy in mucosal melanoma
- PMID: 39513365
- PMCID: PMC11601749
- DOI: 10.1172/jci.insight.179982
Diversity of the immune microenvironment and response to checkpoint inhibitor immunotherapy in mucosal melanoma
Abstract
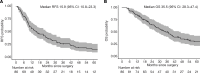
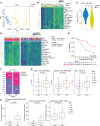
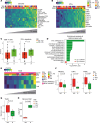
Mucosal melanoma (MucM) is a rare cancer with a poor prognosis and low response rate to immune checkpoint inhibition (ICI) compared with cutaneous melanoma (CM). To explore the immune microenvironment and potential drivers of MucM's relative resistance to ICI drugs, we characterized 101 MucM tumors (43 head and neck [H&N], 31 female urogenital, 13 male urogenital, 11 anorectal, and 3 other gastrointestinal) using bulk RNA-Seq and immunofluorescence. RNA-Seq data show that MucM has a significantly lower IFN-γ signature levels than CM. MucM tumors of the H&N region show a significantly greater abundance of CD8+ T cells, cytotoxic cells, and higher IFN-γ signature levels than MucM from lower body sites. In the subcohort of 35 patients with MucM treated with ICI, hierarchical clustering reveals clusters with a high and low degree of immune infiltration, with a differential ICI response rate. Immune-associated gene sets were enriched in responders. Signatures associated with cancer-associated fibroblasts, macrophages, and TGF-β signaling may be higher in immune-infiltrated, but ICI-unresponsive tumors, suggesting a role for these resistance mechanisms in MucM. Our data show organ region-specific differences in immune infiltration and IFN-γ signature levels in MucM, with H&N MucM displaying the most favorable immune profile. Our study might offer a starting point for developing more personalized treatment strategies for this disease.
Keywords: Cancer immunotherapy; Immunology; Melanoma; Molecular biology; Oncology.
Figures

References
-
- Chang AE, et al. The National Cancer Data Base report on cutaneous and noncutaneous melanoma: a summary of 84,836 cases from the past decade. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer. 1998;83(8):1664–1678. doi: 10.1002/(SICI)1097-0142(19981015)83:8<1664::AID-CNCR23>3.0.CO;2-G. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

