Epigenetic training of human bronchial epithelium cells by repeated rhinovirus infections
- PMID: 39513674
- PMCID: PMC11657021
- DOI: 10.1111/all.16388
Epigenetic training of human bronchial epithelium cells by repeated rhinovirus infections
Abstract
Background: Humans are subjected to various environmental stressors (bacteria, viruses, pollution) throughout life. As such, an inherent relationship exists between the effect of these exposures with age. The impact of these environmental stressors can manifest through DNA methylation (DNAm). However, whether these epigenetic effects selectively target genes, pathways, and biological regulatory mechanisms remains unclear. Due to the frequency of human rhinovirus (HRV) infections throughout life (particularly in early development), we propose the use of HRV under controlled conditions can model the effect of multiple exposures to environmental stressors.
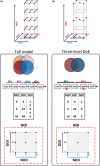
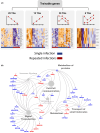
Methods: We generated a prediction model by combining transcriptome and DNAm datasets from human epithelial cells after repeated HRV infections. We applied a novel experimental statistical design and method to systematically explore the multifaceted experimental space (number of infections, multiplicity of infections and duration). Our model included 35 samples, each characterized by the three parameters defining their infection status.
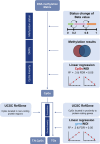
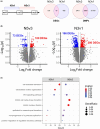
Results: Trainable genes were defined by a consistent linear directionality in DNAm and gene expression changes with successive infections. We identified 77 trainable genes which could be further explored in future studies. The identified methylation sites were tracked within a pediatric cohort to determine the relative changes in candidate-trained sites with disease status and age.
Conclusions: Repeated viral infections induce an immune training response in bronchial epithelial cells. Training-sensitive DNAm sites indicate alternate divergent associations in asthma compared to healthy individuals. Our novel model presents a robust tool for identifying trainable genes, providing a foundation for future studies.
Keywords: airway epithelium; asthma; human rhinovirus; trainable genes; trained immunity.
© 2024 The Author(s). Allergy published by European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
MAR, KDR, SSPN, CJ, CSW, TB, GH, EVM, AMD, RG, NM, FB, SM, and UJ declare no conflict of interest regarding the content of this manuscript. MW reports grants from the COVID‐19 Research Initiative Schleswig‐Holstein, the Follow‐Up of Respiratory Infections in Schleswig‐Holstein (FRISH), the German Center of Lung Research (DZL, Funding No. 82DZL001B6), intramural funding of the Christian‐Albrechts‐University Kiel, the University of Lübeck, and the Leibniz Lung Center, Research Center Borstel. Funding institutions did not participate in the design and conduct of this study. MVK reports grant from the BMBF for the Deutsches Zentrum für Lungenforschung (DZL) and received consultant fees from Sanofi Aventis GmbH, Chiesi GmbH, Allergopharma GmbH along with payments or honoraria for lectures, presentations, speakers' bureaus, manuscript writing, or educational events from Sanofi Aventis GmbH, Infectopharm GmbH, Allergopharma GmbH. KFR received personal payments or honoraria from AstraZeneca, Boehringer Ingelheim, Chiesi Pharmaceuticals, CSL Behring, Sanofi & Regeneron, GlaxoSmithKlfvine, Berlin Chemie, and Menarini; K.F. Rabe also discloses participation on data safety monitoring boards/advisory boards for AstraZeneca, Boehringer Ingelheim, and Sanofi Regeneron, and leadership or fiduciary role in the German Center for Lung Research (DZL), German Chest Society (DGP), and American Thoracic Society (ATS). BS reports grants from the BMBF (German center for lung research, CPC‐Munich, DZL 82DZL033C2, Combat Lung diseases FP4), the German Center for Child and Adolescent Health (DZKJ; LMU/LMU Klinikum: 01GL2406A), from DFG (DFG‐SCHA 997/8–1 (BS); DFG‐SCHA 997/9–1, DFG‐SCHA‐997/10–1, DFG‐SCHA‐997/11–1). BS reports consulting fees from GlaxoSmithKline, Novartis, and Sanofi; payment/honoraria and participation on a Data Safety Monitoring Board or Advisory Board from Sanofi.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

