A Tumor-Naïve ctDNA Assay Detects Minimal Residual Disease in Resected Stage II or III Colorectal Cancer and Predicts Recurrence: Subset Analysis from the GALAXY Study in CIRCULATE-Japan
- PMID: 39513962
- PMCID: PMC11739780
- DOI: 10.1158/1078-0432.CCR-24-2396
A Tumor-Naïve ctDNA Assay Detects Minimal Residual Disease in Resected Stage II or III Colorectal Cancer and Predicts Recurrence: Subset Analysis from the GALAXY Study in CIRCULATE-Japan
Abstract
Purpose: Analysis of ctDNA may enable early identification of patients likely to relapse, presenting an opportunity for early interventions and improved outcomes. Tumor-naïve plasma-only approaches for minimal residual disease (MRD) assessment accelerate turnaround time, enabling rapid treatment decisions and ongoing surveillance.
Experimental design: Plasma samples were obtained from 80 study participants with stage II or III colorectal cancer selected from CIRCULATE-Japan GALAXY. MRD status was assessed using a tumor-naïve ctDNA assay (xM) that integrates methylation and genomic variant data, delivering a binary call. MRD was assessed at 4 weeks postsurgery [landmark time point (LMT)] using methylation and genomic variant data and longitudinally (median, 22.1 months) using only methylation data.
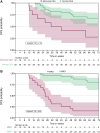
Results: At LMT, 69/80 study participants were evaluable (36 recurrent; 33 nonrecurrent). Of recurrent study participants, 22/36 had detectable ctDNA (MRD-positive) at LMT and 29/33 nonrecurrent study participants had undetectable ctDNA (MRD-negative), yielding a clinical sensitivity of 61.1% and specificity of 87.9%. Additionally, 74 study participants were evaluable for longitudinal performance with a clinical sensitivity of 83.3% and specificity of 89.5%. The median lead time from the first MRD-positive result to recurrence was 4.77 months overall, and 5.30 months for study participants with no adjuvant treatment. At 12 weeks postsurgery, MRD status strongly correlated with disease-free survival (adjusted HR, 9.69), outperforming carcinoembryonic antigen correlation (HR, 2.13).
Conclusions: This tumor-naïve MRD assay demonstrated clinically meaningful performance at LMT and longitudinally, accurately predicting clinical recurrence. MRD status was a stronger prognostic biomarker for disease-free survival compared with standard-of-care carcinoembryonic antigen.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
Y. Nakamura reports grants from Tempus AI, Inc. during the conduct of the study, as well as personal fees from Guardant Health Pte Ltd., Natera, Inc., Roche Ltd., Seagen, Inc., Premo Partners, Inc., Takeda Pharmaceutical Co., Ltd., Exact Sciences Corporation, Gilead Sciences, Inc., MSD K.K., and Eisai Co., Ltd.; personal fees from Zeria Pharmaceutical Co., Ltd., Miyarisan Pharmaceutical Co., Ltd., Merck Biopharma Co., Ltd., CareNet, Inc., Hisamitsu Pharmaceutical Co., Inc., Taiho Pharmaceutical Co., Ltd., Becton, Dickinson and Company, and Guardant Health Japan Corp.; personal fees and nonfinancial support from Daiichi Sankyo Co., Ltd., and Chugai Pharmaceutical Co., Ltd.; and grants from Seagen, Inc., Genomedia Inc., Guardant Health AMEA, Inc., Guardant Health, Inc., and Roche Diagnostics K.K. outside the submitted work. K. Kaneva reports personal fees and other support from Tempus AI, Inc.; other support from Tempus AI, Inc. during the conduct of the study; personal fees and other support from Tempus AI, Inc.; and other support from Tempus AI, Inc. outside the submitted work. C. Lo reports personal fees and other support from Tempus AI, Inc., other support from Tempus AI, Inc. outside the submitted work, and a patent for 18/913,883 pending. D. Neems reports personal fees and other support from Tempus AI, Inc., other support from Tempus AI, Inc. outside the submitted work, and a patent for 18/913,883 pending. J.E. Freaney reports personal fees and other support from Tempus AI, Inc. and other support from Tempus AI, Inc. outside the submitted work. S.W. Hyun reports personal fees from Tempus AI, Inc. and Tempus AI, Inc. outside the submitted work. F. Islam reports personal fees from Tempus AI, Inc. and other support from Tempus AI, Inc. outside the submitted work. J. Yamada-Hanff reports personal fees and other support from Tempus AI, Inc., other support from Tempus AI, Inc. during the conduct of the study, and a patent for 18/913,883 pending. T.M. Driessen reports personal fees from Tempus AI, Inc., other support from Tempus AI, Inc. outside the submitted work, and a patent for 18/913,883 pending. A. Sonnenschein reports personal fees from Tempus AI, Inc., other support from Tempus AI, Inc. outside the submitted work, and a patent for 18/913,883 pending. D.F. DeSantis reports personal fees from Tempus AI, Inc. and other support from Tempus AI, Inc. outside the submitted work. J. Watanabe reports grants and personal fees from Medtronic, personal fees from Johnson and Johnson, Eli Lilly and Company, and Takeda, and grants from AMCO, TERUMO, and Stryker Japan outside the submitted work. M. Kotaka reports personal fees from Chugai and Eli Lilly and Company outside the submitted work. S. Mishima reports personal fees from Taiho Pharmaceutical, Chugai Pharmaceutical, and Eli Lilly and Company outside the submitted work. H. Bando reports personal fees from Ono Pharmaceutical, Taiho Pharmaceutical, and Eli Lilly Japan outside the submitted work. K. Yamazaki reports personal fees from Chugai Pharma, Takeda, Yakult, Taiho, Daiichi Sankyo, Merck Biopharma, Eli Lilly and Company, Ono Pharmaceutical, MSD, and Bristol Myers and Squibb outside the submitted work. H. Taniguchi reports personal fees from Chugai Pharmaceutical, Ono Pharmaceutical, and Eli Lilly and Company; grants and personal fees from Takeda; and grants from Daiichi Sankyo outside the submitted work. T. Kato reports other support from Chugai Pharmaceutical Co., Ltd, Takeda Pharmaceutical Company Limited, Ono Pharmaceutical Co., Eli Lilly and Company, and Taiho Pharmaceutical outside the submitted work. C. Sangli reports personal fees from Tempus AI outside the submitted work. R. Tell reports personal fees and other support from Tempus AI, other support from Tempus AI outside the submitted work, and a patent for 18/913,883 pending. R. Blidner reports personal fees from Tempus during the conduct of the study and personal fees from Tempus outside the submitted work. T. Yoshino reports grants from Amgen K.K., Bristol Myers Squibb K.K., Daiichi Sankyo Co., Ltd., Eisai Co., Ltd., FALCO Biosystems Ltd., Genomedia Inc., Medical & Biological Laboratories, Co., Ltd., Merus N.V., Molecular Health GmbH, Nippon Boehringer Ingelheim Co., Ltd., Pfizer Japan Inc., Roche Diagnostics K.K., Sanofi K.K., Sysmex Corp., Taiho Pharmaceutical Co., Ltd.; grants and personal fees from Chugai Pharmaceutical Co., Ltd., MSD K.K., Takeda Pharmaceutical Co., Ltd., Ono Pharmaceutical, and Co., Ltd.; and personal fees from Merck Biopharma Co., Ltd., and Bayer Yakuhin, Ltd. outside the submitted work. K. Sasser reports other support from Tempus outside the submitted work. E. Oki reports other support from Chugai Pharmaceutical, Ono Pharmaceutical, Eli Lilly and Company, Bristol Myers Squibb, and Takeda Pharmaceutical outside the submitted work. H. Nimeiri reports other support from Tempus AI, AbbVie, and Northwestern Medicine outside the submitted work. No disclosures were reported by the other authors.
Figures



References
-
- Böckelman C, Engelmann BE, Kaprio T, Hansen TF, Glimelius B. Risk of recurrence in patients with colon cancer stage II and III: a systematic review and meta-analysis of recent literature. Acta Oncol 2015;54:5–16. - PubMed
-
- André T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, et al. . Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med 2004;350:2343–51. - PubMed
-
- André T, de Gramont A, Vernerey D, Chibaudel B, Bonnetain F, Tijeras-Raballand A, et al. . Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J Clin Oncol 2015;33:4176–87. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

