Lamination-based organoid spatially resolved transcriptomics technique for primary lung and liver organoid characterization
- PMID: 39514315
- PMCID: PMC11573637
- DOI: 10.1073/pnas.2408939121
Lamination-based organoid spatially resolved transcriptomics technique for primary lung and liver organoid characterization
Abstract
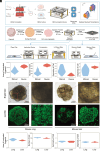
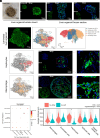
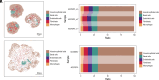
Spatial-transcriptomics technologies have demonstrated exceptional performance in characterizing brain and visceral organ tissues, as well as brain and retinal organoids. However, it has not yet been proven whether spatial transcriptomics can effectively characterize primary tissue-derived organoids, as the standardized tissue sectioning or slicing methods are not applicable for such organoids. Herein, we present a technique, lamination-based organoid spatially resolved transcriptomics (LOSRT), for organoid-spatially resolved transcriptomics based on organoid lamination. Primary mouse lung and liver-derived organoids were used in this study. The organoids were formulated using the droplet-engineering method and laminated using a homemade device with weight compression. This technique preserved most cells in individual organoids while maintaining delicate epithelium structures in laminated domains that can be recognized through visual segmentation. The mouse lung and liver organoids were resolved comprising various cell types, including alveolar cells, damage-associated transient progenitor cells, basal cells, macrophages, endothelial cells, fibroblasts, hepatocytes, and hepatic stellate cells. The distribution and count of cells were confirmed using immunohistology and identified with spatial transcriptomic features. This study reports an automated and integrated spatial transcriptomics method for primary organoids. It has the potential to standardize and rapidly characterize primary tissue-derived organoids.
Keywords: organoid; organoid lamination; spatial transcriptomics.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
-
- Polak R., Zhang E. T., Kuo C. J., Cancer organoids 2.0: Modelling the complexity of the tumour immune microenvironment. Nat. Rev. Cancer 24, 523–539 (2024). - PubMed
-
- Chrisnandy A., Blondel D., Rezakhani S., Broguiere N., Lutolf M. P., Synthetic dynamic hydrogels promote degradation-independent in vitro organogenesis. Nat. Mater. 21, 479–487 (2022). - PubMed
-
- Chen A., et al. , Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792.e21 (2022). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

