Protocol for isolating specific C. elegans neuron types for bulk and single-cell RNA sequencing
- PMID: 39514392
- PMCID: PMC11574799
- DOI: 10.1016/j.xpro.2024.103439
Protocol for isolating specific C. elegans neuron types for bulk and single-cell RNA sequencing
Abstract
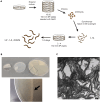
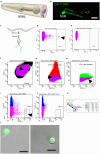
The C. elegans nervous system is compact and well described, ideal for identifying genetic programs that drive neuron-specific development, function, and connectivity. Here, we present a protocol for isolating specific neuron types for gene expression profiling by bulk or single-cell RNA sequencing. We describe steps for worm synchronization, dissociation, and fluorescence-activated cell sorting (FACS) isolation. We then detail procedures for RNA extraction and preparing cells for single-cell sequencing. This protocol is applicable to the isolation of individual cell types from larval and adult animals. For additional details on the use and execution of this protocol, see Taylor et al.1.
Keywords: cell biology; genomics; model organisms; neuroscience.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

