A deep learning model for prediction of autism status using whole-exome sequencing data
- PMID: 39514604
- PMCID: PMC11578481
- DOI: 10.1371/journal.pcbi.1012468
A deep learning model for prediction of autism status using whole-exome sequencing data
Abstract
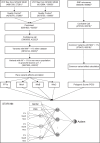
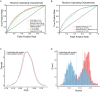
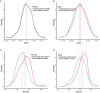
Autism is a developmental disability. Research demonstrated that children with autism benefit from early diagnosis and early intervention. Genetic factors are considered major contributors to the development of autism. Machine learning (ML), including deep learning (DL), has been evaluated in phenotype prediction, but this method has been limited in its application to autism. We developed a DL model, the Separate Translated Autism Research Neural Network (STAR-NN) model to predict autism status. The model was trained and tested using whole exome sequencing data from 43,203 individuals (16,809 individuals with autism and 26,394 non-autistic controls). Polygenic scores from common variants and the aggregated count of rare variants on genes were used as input. In STAR-NN, protein truncating variants, possibly damaging missense variants and mild effect missense variants on the same gene were separated at the input level and merged to one gene node. In this way, rare variants with different level of pathogenic effects were treated separately. We further validated the performance of STAR-NN using an independent dataset, including 13,827 individuals with autism and 14,052 non-autistic controls. STAR-NN achieved a modest ROC-AUC of 0.7319 on the testing dataset and 0.7302 on the independent dataset. STAR-NN outperformed other traditional ML models. Gene Ontology analysis on the selected gene features showed an enrichment for potentially informative pathways including calcium ion transport.
Copyright: © 2024 Wu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
No competing interests.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

