Macrophage LRRK2 hyperactivity impairs autophagy and induces Paneth cell dysfunction
- PMID: 39514635
- PMCID: PMC11730131
- DOI: 10.1126/sciimmunol.adi7907
Macrophage LRRK2 hyperactivity impairs autophagy and induces Paneth cell dysfunction
Abstract
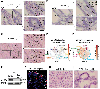
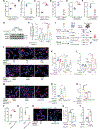
LRRK2 polymorphisms (G2019S/N2081D) that increase susceptibility to Parkinson's disease and Crohn's disease (CD) lead to LRRK2 kinase hyperactivity and suppress autophagy. This connection suggests that LRRK2 kinase inhibition, a therapeutic strategy being explored for Parkinson's disease, may also benefit patients with CD. Paneth cell homeostasis is tightly regulated by autophagy, and their dysfunction is a precursor to gut inflammation in CD. Here, we found that patients with CD and mice carrying hyperactive LRRK2 polymorphisms developed Paneth cell dysfunction. We also found that LRRK2 kinase can be activated in the context of interactions between genes (genetic autophagy deficiency) and the environment (cigarette smoking). Unexpectedly, lamina propria immune cells were the main intestinal cell types that express LRRK2, instead of Paneth cells as previously suggested. We showed that LRRK2-mediated pro-inflammatory cytokine release from phagocytes impaired Paneth cell function, which was rescued by LRRK2 kinase inhibition through activation of autophagy. Together, these data suggest that LRRK2 kinase inhibitors maintain Paneth cell homeostasis by restoring autophagy and may represent a therapeutic strategy for CD.
Conflict of interest statement
Figures




References
-
- Thaiss CA, Zmora N, Levy M, Elinav E, The microbiome and innate immunity. Nature 535, 65–74 (2016). - PubMed
-
- Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Böck J, Martinez-Naves E, Glickman JN, Tschurtschenthaler M, Hartwig J, Hosomi S, Flak MB, Cusick JL, Kohno K, Iwawaki T, Billmann-Born S, Raine T, Bharti R, Lucius R, Kweon MN, Marciniak SJ, Choi A, Hagen SJ, Schreiber S, Rosenstiel P, Kaser A, Blumberg RS, Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276 (2013). - PMC - PubMed
-
- Clevers HC, Bevins CL, Paneth cells: Maestros of the small intestinal crypts. Annu. Rev. Physiol. 75, 289–311 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

